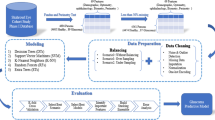
Abstract
In managing patients with chronic diseases, such as open angle glaucoma (OAG), the case treated in this paper, medical tests capture the disease phase (e.g. regression, stability, progression, etc.) the patient is currently in. When medical tests have low residual variability (e.g. empirical difference between the patient’s true and recorded value is small) they can effectively, without the use of sophisticated methods, identify the patient’s current disease phase; however, when medical tests have moderate to high residual variability this may not be the case. This paper presents a framework for handling the latter case. The framework presented integrates the outputs of interacting multiple model Kalman filtering with supervised learning classification. The purpose of this integration is to estimate the true values of patients’ disease metrics by allowing for rapid and non-rapid phases; and dynamically adapting to changes in these values over time. We apply our framework to classifying whether a patient with OAG will experience rapid progression over the next two or three years from the time of classification. The performance (AUC) of our model increased by approximately 7% (increased from 0.752 to 0.819) when the Kalman filtering results were incorporated as additional features in the supervised learning model. These results suggest the combination of filters and statistical learning methods in clinical health has significant benefits. Although this paper applies our methodology to OAG, the methodology developed is applicable to other chronic conditions.



Similar content being viewed by others
References
(2020) National center for chronic disease prevention and health promotion. https://www.cdc.gov/chronicdisease/index.htm
Amaral JL, Lopes AJ, Jansen JM, Faria AC, Melo PL (2012) Machine learning algorithms and forced oscillation measurements applied to the automatic identification of chronic obstructive pulmonary disease. Comput Methods Progr Biomed 105(3):183–193
Bergmeir C, Benítez JM (2012) On the use of cross-validation for time series predictor evaluation. Inf Sci 191:192–213
Blom HA (1984) An efficient filter for abruptly changing systems. In: The 23rd IEEE conference on decision and control. IEEE, pp 656–658
Blom HA, Bar-Shalom Y (1988) The interacting multiple model algorithm for systems with markovian switching coefficients. IEEE Trans Autom Control 33(8):780–783
Brodersen KH, Ong CS, Stephan KE, Buhmann JM (2010) The balanced accuracy and its posterior distribution. In: 2010 20th International conference on pattern recognition. IEEE, pp 3121–3124
Catal C, Nangir M (2017) A sentiment classification model based on multiple classifiers. Appl Soft Comput 50:135–141
Corbin JM (1991) A nursing model for chronic illness management based upon the trajectory framework. Res Theory Nurs Pract 5(3):155
Das A, Rad P, Choo KKR, Nouhi B, Lish J, Martel J (2019) Distributed machine learning cloud teleophthalmology iot for predicting amd disease progression. Future Gener Comput Syst 93:486–498
Dietterich TG (2000) Ensemble methods in machine learning. In: International workshop on multiple classifier systems. Springer, pp 1–15
Friedman J, Hastie T, Tibshirani R (2001) The elements of statistical learning, vol 1, Springer Series in Statistics, New York
Garcia GGP, Lavieri MS, Andrews C, Liu X, Van Oyen MP, Kass MA, Gordon MO, Stein JD (2019a) Accuracy of Kalman filtering in forecasting visual field and intraocular pressure trajectory in patients with ocular hypertension. JAMA Ophthalmol 137(12):1416–1423
Garcia GGP, Nitta K, Lavieri MS, Andrews C, Liu X, Lobaza E, Van Oyen MP, Sugiyama K, Stein JD (2019b) Using Kalman filtering to forecast disease trajectory for patients with normal tension glaucoma. Am J Ophthalmol 199:111–119
Géron A (2017) Hands-on machine learning with Scikit-Learn and TensorFlow: concepts, tools and techniques to build intelligent systems. O’Reilly Media, Inc
Gracitelli CP, Zangwill LM, Diniz-Filho A, Abe RY, Girkin CA, Weinreb RN, Liebmann JM, Medeiros FA (2018) Detection of glaucoma progression in individuals of african descent compared with those of european descent. JAMA Ophthalmol 136(4):329–335
Group EDPR, et al. (2004) Prevalence of open-angle glaucoma among adults in the united states. Arch Ophthalmol 122(4):532
Heijl A, Leske MC, Bengtsson B, Bengtsson B, Hussein M, Group E (2003) Measuring visual field progression in the early manifest glaucoma trial. Acta Ophthalmol Scand 81(3):286–293
Hutter F, Xu L, Hoos HH, Leyton-Brown K (2014) Algorithm runtime prediction: methods & evaluation. Artif Intell 206:79– 111
Kazemian P, Helm JE, Lavieri MS, Stein JD, Van Oyen MP (2019) Dynamic monitoring and control of irreversible chronic diseases with application to glaucoma. Prod Oper Manag 28(5):1082–1107
Kleiman RS, LaRose ER, Badger JC, Page D, Caldwell MD, Clay JA, Peissig PL (2018) Using machine learning algorithms to predict risk for development of calciphylaxis in patients with chronic kidney disease. In: AMIA Summits on translational science proceedings, p 139
Labbe R (2015) Kalman and Bayesian filters in python. Book
Russell RA, Crabb DP, Malik R, Garway-Heath DF (2012) The relationship between variability and sensitivity in large-scale longitudinal visual field data. Investig Ophthalmol Vis Sci 53(10):5985–5990
Russell RA, Garway-Heath DF, Crabb DP (2013) New insights into measurement variability in glaucomatous visual fields from computer modelling. PloS One 8(12):e83595
Schell GJ, Lavieri MS, Helm JE, Liu X, Musch DC, Van Oyen MP, Stein JD (2014) Using filtered forecasting techniques to determine personalized monitoring schedules for patients with open-angle glaucoma. Ophthalmology 121(8):1539–1546
Stein RM (2002) Benchmarking default prediction models: pitfalls and remedies in model validation. Moody’s KMV, New York, p 20305
Vesti E, Johnson CA, Chauhan BC (2003) Comparison of different methods for detecting glaucomatous visual field progression. Investig Ophthalmol Vis Sci 44(9):3873–3879
Xiao J, Ding R, Xu X, Guan H, Feng X, Sun T, Zhu S, Ye Z (2019) Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J Transl Med 17(1):119
Ye C, Fu T, Hao S, Zhang Y, Wang O, Jin B, Xia M, Liu M, Zhou X, Wu Q et al (2018) Prediction of incident hypertension within the next year: prospective study using statewide electronic health records and machine learning. J Med Internet Res 20(1):e22
Yousefi S, Elze T, Pasquale LR, Boland M (2018) Glaucoma monitoring using manifold learning and unsupervised clustering. In: 2018 International conference on image and vision computing New Zealand (IVCNZ). IEEE, pp 1–6
Zarchan P, Musoff H (2013) Fundamentals of Kalman filtering: a practical approach. American Institute of Aeronautics and Astronautics, Inc
Zhou Q, Zhang Z, Wang Y (2019) Wit120 data mining technology based on internet of things. Health Care Management Science 1–9
Acknowledgements
Supported by the National Eye Institute, NIH R01 EY026641; the National Science Foundation, NSF grant CMMI- 1552525; and the Lighthouse Guild.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Statement
Ethics approval for this work was obtained via University of Michigan IRB HUM00079342.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Partial funding was received from the National Eye - NIH grant R01EY026641 and from a University of Michigan Merit Fellowship
Appendices
Appendix A: Detailed description of case study model inputs
Featured measured | Feature engineering | Description | Data used |
|---|---|---|---|
function | |||
Progression | Expanding window | Using an expanding window measure progression | MD, KF MD |
from periods 3 to T, engineer the features OLSR | |||
slope, OLSR slope p-value, and a binary indicator | |||
indicating if the slope is statistically significant at | |||
the level of 0.05 | |||
Calculate summary statistics of mean, standard | |||
deviation, quantiles at various levels, min, and | |||
max for each covariate described in the above row. | |||
Moving window | Calculate a moving window of 3 most recent | ||
periods from periods 3 to T for OLSR slope, and | |||
OLSR p-value. | |||
Patient Test | Expanding window | Using an expanding window calculate summary | MD, PSD, IOP, |
Results | statistics mean, standard deviation, various | KF MD | |
quantiles, min, and max from periods 0 to T. | |||
Moving window | Using a moving window of 3 & 4 periods, | ||
calculate summary statistics (e.g., mean, standard | |||
deviation, quantiles, min, and max). | |||
Shifted/ Time-lag | Time lag features in addition to time T: T-3, | ||
features | T-2, T-1. | ||
Demographics | N/A | Unchanging covariates based on patient’s | |
demographic information. | |||
Age, Sex, Race | |||
Patient Appointment | N/A | Follow-up period. | Visit Period (6-month |
Information | interval) |
Appendix B: IMM initial filter model parameters
The bank of Kalman Filters was iteratively tuned using a grid search procedure. The objective of this procedure was to determine the best parameters for the Kalman filter models. The initial estimates for M and μ0 were obtained using the data. M was estimated using the number of transitions between RP and RP; RP and Non-RP; Non-RP and RP; and Non-RP and Non-RP. μ0 was estimated using the normalized frequency of RP instances and Non-RP instances. The estimated values for M and μ0 are found in Tables 8 and 9 respectively.
The elements of the F matrix are shown in Table 10. The F matrix captures the system transition of Kalman Filter. The F matrix was built using the linear vector difference equation model for the following physics equations:
where Δt = 6 months, and state variables x = 〈MD MDvelocity MDacceleration〉.
The Q (process) noise covariance matrices are shown in Tables 11 and 12. The Q matrix captures the noise introduced into our system due to external factors we do not directly model for. The Q matrix captures this noise using a random process centered at 0. In our model we assumed the Q matrices have a piecewise white noise model, where the noise follows a discrete time Wiener process. This noise model assumes the noise for the highest order term (e.g. acceleration) is constant for the duration of each time period, but differs for each time period, and each of these is uncorrelated between time periods [21].
The initial estimates for the MD, MD velocity, and MD acceleration variances are high, so as to model the uncertainty associated with these three measurements when you see a new patient (Table 13).
As a clinician with a new patient there is no current information to suggest an appropriate MD rate of change. There is only a single MD baseline value. As such the initial state estimate for each patient is
Last, the measurement function, H, and measurement noise, R are defined respectively as,
Appendix C: Illustration of the IMM Kalman filtered MD results compared to the patient’s MD results and the MD “true state” estimated by OLSR
Rights and permissions
About this article
Cite this article
Jones, I.A., Van Oyen, M. ., Lavieri, M.S. et al. Predicting rapid progression phases in glaucoma using a soft voting ensemble classifier exploiting Kalman filtering. Health Care Manag Sci 24, 686–701 (2021). https://doi.org/10.1007/s10729-021-09564-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10729-021-09564-2