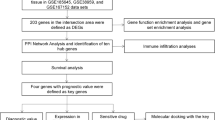
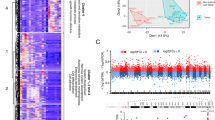
Summary
Neuroblastoma (NB) is a common tumor in children, usually in the retroperitoneum. After various treatments, low- and intermediate-risk patients have achieved good results, but the prognosis of high-risk patients is still very poor. Therefore, it is necessary to find new effective targets for the treatment of high-risk patients. In this study, comprehensive bioinformatics analysis was used to identify the differentially expressed genes (DEG and DEM) between high-risk patients and non-high-risk patients, and it was identified that ADRB2 may affect the survival status of high-risk patients due to miR -30a-5p regulation. The GSE49710, GSE73517, and GSE121513 datasets were downloaded from the Gene Expression Synthesis (GEO) database, and DEG and DEM were selected. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were applied to the selected DEGs. The STRING database and Cytoscape software were used to construct protein-protein interaction (PPI) networks and perform modular analysis of the DEGs. The TARGET data set containing information on overall survival days were used for the prognostic analysis of central genes. We identified a total of 255 DEGs from GSE49710 and GSE73517, and 193 DEMs from GSE121513. We identified the 5 most important central genes from the PPI network, performed a prognostic analysis in the target data set, and verified their expression using RT-qPCR to select the most important ADRB2 gene to predict miRNA. Integrating the differential miRNA predicted by miRDB and miRSystem and GSE121513 between the targeted miRNA and the prognosis, miR-30a-5p was finally identified as the targeted miRNA of ADRB2.







Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and Therapeutically Applicable Research To Generate Effective Treatments (https://ocg.cancer.gov/programs/target) repositories.
References
Kholodenko IV, Kalinovsky DV, Doronin II, Deyev SM, Kholodenko RV (2018) Neuroblastoma origin and therapeutic targets for immunotherapy. J Immunol Res 2018:7394268. https://doi.org/10.1155/2018/7394268
Maris JM, Hogarty MD, Bagatell R, Cohn SL (2007) Neuroblastoma. Lancet 369(9579):2106–2120. https://doi.org/10.1016/S0140-6736(07)60983-0
Fonseka P, Liem M, Ozcitti C, Adda CG, Ang CS, Mathivanan S (2019) Exosomes from N-Myc amplified neuroblastoma cells induce migration and confer chemoresistance to non-N-Myc amplified cells: implications of intra-tumor heterogeneity. J Extracell Vesicles 8(1):1597614. https://doi.org/10.1080/20013078.2019.1597614
Koneru B, Lopez G, Farooqi A, Conkrite KL, Nguyen TH, Macha SJ, Modi A, Rokita JL, Urias E, Hindle A, Davidson H, McCoy K, Nance J, Yazdani V, Irwin MS, Yang S, Wheeler DA, Maris JM, Diskin SJ, Reynolds CP (2020) Telomere maintenance mechanisms define clinical outcome in high-risk neuroblastoma. Cancer Res 80(12):2663–2675. https://doi.org/10.1158/0008-5472.CAN-19-3068
Strother DR, London WB, Schmidt ML, Brodeur GM, Shimada H, Thorner P, Collins MH, Tagge E, Adkins S, Reynolds CP, Murray K, Lavey RS, Matthay KK, Castleberry R, Maris JM, Cohn SL (2012) Outcome after surgery alone or with restricted use of chemotherapy for patients with low-risk neuroblastoma: results of Children's Oncology group study P9641. J Clin Oncol 30(15):1842–1848. https://doi.org/10.1200/JCO.2011.37.9990
Baker DL, Schmidt ML, Cohn SL, Maris JM, London WB, Buxton A, Stram D, Castleberry RP, Shimada H, Sandler A, Shamberger RC, Look AT, Reynolds CP, Seeger RC, Matthay KK, Children's Oncology G (2010) Outcome after reduced chemotherapy for intermediate-risk neuroblastoma. N Engl J Med 363(14):1313–1323. https://doi.org/10.1056/NEJMoa1001527
Pearson AD, Pinkerton CR, Lewis IJ, Imeson J, Ellershaw C, Machin D, European Neuroblastoma Study G, Children's C, Leukaemia G (2008) High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: a randomized trial. Lancet Oncol 9(3):247–256. https://doi.org/10.1016/S1470-2045(08)70069-X
Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, Gerbing RB, London WB, Villablanca JG (2009) Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children's oncology group study. J Clin Oncol 27(7):1007–1013. https://doi.org/10.1200/JCO.2007.13.8925
Holmes K, Potschger U, ADJ P, Sarnacki S, Cecchetto G, Gomez-Chacon J, Squire R, Freud E, Bysiek A, Matthyssens LE, Metzelder M, Monclair T, Stenman J, Rygl M, Rasmussen L, Joseph JM, Irtan S, Avanzini S, Godzinski J, Bjornland K, Elliott M, Luksch R, Castel V, Ash S, Balwierz W, Laureys G, Ruud E, Papadakis V, Malis J, Owens C, Schroeder H, Beck-Popovic M, Trahair T, Forjaz de Lacerda A, Ambros PF, Gaze MN, McHugh K, Valteau-Couanet D, Ladenstein RL, International Society of Paediatric Oncology Europe Neuroblastoma G (2020) Influence of surgical excision on the survival of patients with stage 4 high-risk neuroblastoma: a report from the HR-NBL1/SIOPEN study. J Clin Oncol 38(25):2902–2915. https://doi.org/10.1200/JCO.19.03117
Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, Valteau-Couanet D, Pearson AD, Cohn SL (2015) Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol 33(27):3008–3017. https://doi.org/10.1200/JCO.2014.59.4648
Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA (2016) Neuroblastoma. Nat Rev Dis Primers 2:16078. https://doi.org/10.1038/nrdp.2016.78
Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, Hogarty M, Committee COGN (2013) Children's Oncology Group's 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer 60(6):985–993. https://doi.org/10.1002/pbc.24433
Pstrag N, Ziemnicka K, Bluyssen H, Wesoly J (2018) Thyroid cancers of follicular origin in a genomic light: in-depth overview of common and unique molecular marker candidates. Mol Cancer 17(1):116. https://doi.org/10.1186/s12943-018-0866-1
Toro-Dominguez D, Martorell-Marugan J, Lopez-Dominguez R, Garcia-Moreno A, Gonzalez-Rumayor V, Alarcon-Riquelme ME, Carmona-Saez P (2019) ImaGEO: integrative gene expression meta-analysis from GEO database. Bioinformatics 35(5):880–882. https://doi.org/10.1093/bioinformatics/bty721
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R (2007) NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res 35(Database issue):D760–D765. https://doi.org/10.1093/nar/gkl887
Chen B, Ding P, Hua Z, Qin X, Li Z (2020) Analysis and identification of novel biomarkers involved in neuroblastoma via integrated bioinformatics. Investig New Drugs. https://doi.org/10.1007/s10637-020-00980-9
Wang H, Wang X, Xu L, Zhang J, Cao H (2020) Prognostic significance of MYCN related genes in pediatric neuroblastoma: a study based on TARGET and GEO datasets. BMC Pediatr 20(1):314. https://doi.org/10.1186/s12887-020-02219-1
Zhong X, Liu Y, Liu H, Zhang Y, Wang L, Zhang H (2018) Identification of potential prognostic genes for neuroblastoma. Front Genet 9:589. https://doi.org/10.3389/fgene.2018.00589
Clough E, Barrett T (2016) The gene expression omnibus database. Methods Mol Biol 1418:93–110. https://doi.org/10.1007/978-1-4939-3578-9_5
Consortium SM-I (2014) A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat Biotechnol 32(9):903–914. https://doi.org/10.1038/nbt.2957
Henrich KO, Bender S, Saadati M, Dreidax D, Gartlgruber M, Shao C, Herrmann C, Wiesenfarth M, Parzonka M, Wehrmann L, Fischer M, Duffy DJ, Bell E, Torkov A, Schmezer P, Plass C, Hofer T, Benner A, Pfister SM, Westermann F (2016) Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas. Cancer Res 76(18):5523–5537. https://doi.org/10.1158/0008-5472.CAN-15-2507
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264. https://doi.org/10.1093/biostatistics/4.2.249
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20(3):307–315. https://doi.org/10.1093/bioinformatics/btg405
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Diboun I, Wernisch L, Orengo CA, Koltzenburg M (2006) Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 7:252. https://doi.org/10.1186/1471-2164-7-252
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34. https://doi.org/10.1093/nar/27.1.29
Sherman BT, Huang d W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) DAVID knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8:426. https://doi.org/10.1186/1471-2105-8-426
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(Database issue):D447–D452. https://doi.org/10.1093/nar/gku1003
Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2019) Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 18(2):623–632. https://doi.org/10.1021/acs.jproteome.8b00702
Park JA, Cheung NV (2020) Targets and antibody formats for immunotherapy of neuroblastoma. J Clin Oncol 38(16):1836–1848. https://doi.org/10.1200/JCO.19.01410
Xia XQ, Jia Z, Porwollik S, Long F, Hoemme C, Ye K, Muller-Tidow C, McClelland M, Wang Y (2010) Evaluating oligonucleotide properties for DNA microarray probe design. Nucleic Acids Res 38(11):e121. https://doi.org/10.1093/nar/gkq039
Pounds S, Morris SW (2003) Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 19(10):1236–1242. https://doi.org/10.1093/bioinformatics/btg148
Kupfer P, Guthke R, Pohlers D, Huber R, Koczan D, Kinne RW (2012) Batch correction of microarray data substantially improves the identification of genes differentially expressed in rheumatoid arthritis and osteoarthritis. BMC Med Genet 5:23. https://doi.org/10.1186/1755-8794-5-23
Karstens KF, Bellon E, Polonski A, Wolters-Eisfeld G, Melling N, Reeh M, Izbicki JR, Tachezy M (2020) Expression and serum levels of the neural cell adhesion molecule L1-like protein (CHL1) in gastrointestinal stroma tumors (GIST) and its prognostic power. Oncotarget 11(13):1131–1140. https://doi.org/10.18632/oncotarget.27525
Svensmark JH, Brakebusch C (2019) Rho GTPases in cancer: friend or foe? Oncogene 38(50):7447–7456. https://doi.org/10.1038/s41388-019-0963-7
Schwab M, Ellison J, Busch M, Rosenau W, Varmus HE, Bishop JM (1984) Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc Natl Acad Sci U S A 81(15):4940–4944. https://doi.org/10.1073/pnas.81.15.4940
Tajbakhsh A, Pasdar A, Rezaee M, Fazeli M, Soleimanpour S, Hassanian SM, FarshchiyanYazdi Z, Younesi Rad T, Ferns GA, Avan A (2018) The current status and perspectives regarding the clinical implication of intracellular calcium in breast cancer. J Cell Physiol 233(8):5623–5641. https://doi.org/10.1002/jcp.26277
Haug BH, Hald OH, Utnes P, Roth SA, Lokke C, Flaegstad T, Einvik C (2015) Exosome-like extracellular vesicles from MYCN-amplified neuroblastoma cells contain oncogenic miRNAs. Anticancer Res 35(5):2521–2530
Ohashi K, Wang Z, Yang YM, Billet S, Tu W, Pimienta M, Cassel SL, Pandol SJ, Lu SC, Sutterwala FS, Bhowmick N, Seki E (2019) NOD-like receptor C4 Inflammasome regulates the growth of Colon Cancer liver metastasis in NAFLD. Hepatology 70(5):1582–1599. https://doi.org/10.1002/hep.30693
Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Nunez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA, O'Neill LA (2015) A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 21(3):248–255. https://doi.org/10.1038/nm.3806
Philipp M, Hein L (2004) Adrenergic receptor knockout mice: distinct functions of 9 receptor subtypes. Pharmacol Ther 101(1):65–74. https://doi.org/10.1016/j.pharmthera.2003.10.004
Sassone-Corsi P (2012) The cyclic AMP pathway. Cold Spring Harb Perspect Biol 4(12). https://doi.org/10.1101/cshperspect.a011148
Sapio L, Gallo M, Illiano M, Chiosi E, Naviglio D, Spina A, Naviglio S (2017) The natural cAMP elevating compound Forskolin in Cancer therapy: is it time? J Cell Physiol 232(5):922–927. https://doi.org/10.1002/jcp.25650
Bergantin LB (2019) Diabetes and cancer: debating the link through ca(2+)/cAMP signalling. Cancer Lett 448:128–131. https://doi.org/10.1016/j.canlet.2019.02.017
Zhang X, Zhang Y, He Z, Yin K, Li B, Zhang L, Xu Z (2019) Chronic stress promotes gastric cancer progression and metastasis: an essential role for ADRB2. Cell Death Dis 10(11):788. https://doi.org/10.1038/s41419-019-2030-2
Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, Maurer HC, Chen X, Jiang Z, Westphalen CB, Ilmer M, Valenti G, Mohanta SK, Habenicht AJR, Middelhoff M, Chu T, Nagar K, Tailor Y, Casadei R, Di Marco M, Kleespies A, Friedman RA, Remotti H, Reichert M, Worthley DL, Neumann J, Werner J, Iuga AC, Olive KP, Wang TC (2018) beta2 adrenergic-Neurotrophin feedforward loop promotes pancreatic Cancer. Cancer Cell 33 (1):75–90 e77. doi:https://doi.org/10.1016/j.ccell.2017.11.007
Thaker PH, Han LY, Kamat AA, Arevalo JM, Takahashi R, Lu C, Jennings NB, Armaiz-Pena G, Bankson JA, Ravoori M, Merritt WM, Lin YG, Mangala LS, Kim TJ, Coleman RL, Landen CN, Li Y, Felix E, Sanguino AM, Newman RA, Lloyd M, Gershenson DM, Kundra V, Lopez-Berestein G, Lutgendorf SK, Cole SW, Sood AK (2006) Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat Med 12(8):939–944. https://doi.org/10.1038/nm1447
Braadland PR, Ramberg H, Grytli HH, Urbanucci A, Nielsen HK, Guldvik IJ, Engedal A, Ketola K, Wang W, Svindland A, Mills IG, Bjartell A, Tasken KA (2019) The beta2-adrenergic receptor is a molecular switch for neuroendocrine Transdifferentiation of prostate Cancer cells. Mol Cancer Res 17(11):2154–2168. https://doi.org/10.1158/1541-7786.MCR-18-0605
Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, Li T, Wang CZ, Tan YX, Ding J, Chen Y, Tang L, Guo LN, Tang SH, Yang W, Wang HY (2016) ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol 65(2):314–324. https://doi.org/10.1016/j.jhep.2016.04.019
Xie WY, He RH, Zhang J, He YJ, Wan Z, Zhou CF, Tang YJ, Li Z, McLeod HL, Liu J (2019) Betablockers inhibit the viability of breast cancer cells by regulating the ERK/COX2 signaling pathway and the drug response is affected by ADRB2 singlenucleotide polymorphisms. Oncol Rep 41(1):341–350. https://doi.org/10.3892/or.2018.6830
Kulik G (2019) ADRB2-targeting therapies for prostate Cancer. Cancers (Basel) 11(3). https://doi.org/10.3390/cancers11030358
Li L, Kang L, Zhao W, Feng Y, Liu W, Wang T, Mai H, Huang J, Chen S, Liang Y, Han J, Xu X, Ye Q (2017) miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett 400:89–98. https://doi.org/10.1016/j.canlet.2017.04.034
Zhou L, Jia S, Ding G, Zhang M, Yu W, Wu Z, Cao L (2019) Down-regulation of miR-30a-5p is associated with poor prognosis and promotes Chemoresistance of gemcitabine in pancreatic ductal adenocarcinoma. J Cancer 10(21):5031–5040. https://doi.org/10.7150/jca.31191
Tao J, Cong H, Wang H, Zhang D, Liu C, Chu H, Qing Q, Wang K (2018) MiR-30a-5p inhibits osteosarcoma cell proliferation and migration by targeting FOXD1. Biochem Biophys Res Commun 503(2):1092–1097. https://doi.org/10.1016/j.bbrc.2018.06.121
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, Hou J, Lin L, Cai J (2018) Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med 16(1):216. https://doi.org/10.1186/s12967-018-1582-8
Yang J, Song H, Cao K, Song J, Zhou J (2019) Comprehensive analysis of helicobacter pylori infection-associated diseases based on miRNA-mRNA interaction network. Brief Bioinform 20(4):1492–1501. https://doi.org/10.1093/bib/bby018
Xue J, Zhou D, Poulsen O, Hartley I, Imamura T, Xie EX, Haddad GG (2018) Exploring miRNA-mRNA regulatory network in cardiac pathology in Na(+)/H(+) exchanger isoform 1 transgenic mice. Physiol Genomics 50(10):846–861. https://doi.org/10.1152/physiolgenomics.00048.2018
Funding
This work was supported by the Key Project of “Research on Prevention and Control of Major Chronic Non-Communicable Diseases”, Ministry of Science and Technology of the People’s Republic of China (2018YFC1313004), Chongqing Science and Technology Commission (cstc2019jscx-msxmX0220); General Clinical Medical Research Program of Children’s Hospital of Chongqing Medical University (YBXM-2019-003).
Author information
Authors and Affiliations
Contributions
Shao performed the research, analyzed the data, and wrote the manuscript, Liu collected the clinical samples, and Wang* contributed to carrying out additional analyses, discussed the results, provided scientific advice, and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The entire study was approved by the Medical Ethics Committee of Children’s Hospital of Chongqing Medical University and complies with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
For this type of study, formal consent is not required.
Consent to participate
Not applicable.
Consent for publication
All authors consent to the publication of this study.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shao, FL., Liu, Qq. & Wang, S. Identify potential miRNA-mRNA regulatory networks contributing to high-risk neuroblastoma. Invest New Drugs 39, 901–913 (2021). https://doi.org/10.1007/s10637-021-01064-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-021-01064-y