Abstract
Background and Aims
The present study aimed to identify the differential expressed genes that are related to acute pancreatitis.
Methods
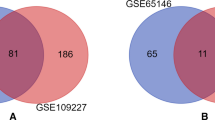
Microarray datasets GSE109227 and GSE3644 were downloaded from the public database and analyzed to screen the genes. Afterward, integrated analysis of these genes were performed, including gene ontology and pathway enrichment analysis, protein–protein interaction network construction, expression level evaluation in human organs, relevant miRNAs and TFs prediction, and prognosis values of hub genes in pancreatic carcinoma.
Results
A total number of 93 differential expressed genes were screened from the datasets, and EGFR, CDH1, ACTB, CD44, and VCL were identified as hub DEGs. Functional enrichment analysis demonstrated that these genes were mostly enriched in biological processes such as cell adhesion, platelet aggregation, glycoprotein binding, and also involved in multiple pathways included adherent junction, proteoglycans in cancer, bacterial invasion of epithelial cells, focal adhesion, Rap1 signaling pathway, regulation of actin cytoskeleton, and pathways in cancers. The five hub genes were all expressed in human pancreas organs with various levels. Hub gene-related network investigation predicted core miRNAs including hsa-mir-16-5p and main TFs like SOX9 with close interactions with these hub genes. Survival analysis also indicated that the high expression of EGFR, CDH1, ACTB, CD44, and VCL were significantly associated with poor prognosis in pancreatic carcinoma.
Conclusions
The study suggested that hub genes EGFR, CDH1, ACTB, CD44, and VCL may play vital role in the pathogenesis of acute pancreatitis and may serve as potential biomarkers to facilitate future acute pancreatitis diagnosis and treatment.







Similar content being viewed by others
References
Forsmark CE, Vege SS, Wilcox CM. Acute pancreatitis. N Engl J Med. 2017;376:598–599.
Venneman NG, Buskens E, Besselink MG, et al. Small gallstones are associated with increased risk of acute pancreatitis: potential benefits of prophylactic cholecystectomy? Am J Gastroenterol. 2005;100:2540–2550.
Lankisch PG, Lowenfels AB, et al. What is the risk of alcoholic pancreatitis in heavy drinkers? Pancreas. 2002;25:411–412.
Banks PA, Bollen TL, Dervenis C, the Acute Pancreatitis Classification Working Group, et al. Classification of acute pancreatitis-2012: revision of the Atlanta classification and definitions by international consensus. Gut. 2013;62:102–111.
Lankisch PG, Apte M, Banks PA. Acute pancreatitis. Lancet. 2015;386:85–96.
Mckay CJ, Imrie CW. The continuing challenge of early mortality in acute pancreatitis. Br J Surg. 2010;91:1243–1244.
Whitcomb David C. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144:1292–1302.
Pelaez-Luna M, Robles-Diaz G, Canizales-Quinteros S, et al. PRSS1 and SPINK1 mutations in idiopathic chronic and recurrent acute pancreatitis. World J Gastroenterol. 2014;20:11788–11792.
Xiao H, Xu D, Chen P, Zeng G, et al. Identification of five genes as a potential biomarker for predicting progress and prognosis in adrenocortical carcinoma. J Cancer.. 2018;9:4484–4495.
Cheok CF, Verma CS, Baselga J, et al. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37.
Lehmann S, Bykov VJN, Ali D, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:561–568.
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2.
Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:6352.
Vlachos IS, Paraskevopoulou D, Karagkouni G, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucl Acids Res. 2015;43:D153–D159.
Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:2017.
Xia J, Gill EE, Hancock REW. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10:823–844.
Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102.
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J PrePrints. 2016;2:e67.
Ozaki N, Fukuchi Y, Tomiyoshi S, et al. Autophagy regulation in pancreatic acinar cells is independent of epidermal growth factor receptor signaling. Biochem Biophys Res Commun. 2014;446:224–230.
Lu M. Construction of network for protein kinases that play a role in acute pancreatitis. Pancreas. 2013;42:607–613.
Huang Y, Xia W, Lu M, et al. Role of kinase epidermal growth factor receptor and SRC in the Caerulein-induced acute pancreatitis in mice. Pancreas. 2014;44:152–157.
Hu G, Shen J, Cheng L, et al. Reg4 protects against acinar cell necrosis in experimental pancreatitis. Gut. 2011;60:820.
Dembiński A, Warzecha Z, Konturek PC, et al. Epidermal growth factor accelerates pancreatic recovery after caerulein-induced pancreatitis. Eur J Pharmacol. 2000;398:159–168.
Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634.
Serrill JD, Sander M, Shih HP. Pancreatic exocrine tissue architecture and integrity are maintained by e-cadherin during postnatal development. Sci Rep. 2018;8:13451.
Mayerle J, Schnekenburger J, Kruger B, et al. Extracellular cleavage of E-cadherin by leukocyte elastase during acute experimental pancreatitis in rats. Gastroenterology. 2005;129:1251–1267.
Virtanen JA, Vartiainen MK. Diverse functions for different forms of nuclear actin. Curr Opin Cell Biol. 2017;46:33.
Gunning PW, Ghoshdastider U, Whitaker S, et al. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128:2009–2019.
Yuan S, Rosenberg L, Ilieva A, et al. Early changes of gene expression during cerulein supramaximal stimulation. Pancreas. 1999;19:45.
Yan Z, Gao J, Lv X, et al. Quantitative evaluation and selection of reference genes for quantitative RT-PCR in mouse acute pancreatitis. Biomed Res Int. 2016;2016:2016.
Naor D. CD44: structure, function, and association with the malignant process. Adv Cancer Res.. 1997;71:241–319.
Durko L, Wlodarski W, Stasikowskakanicka O, et al. Expression and clinical significance of cancer stem cell markers CD24, CD44, and CD133 in pancreatic ductal adenocarcinoma and chronic pancreatitis. Dis Mark. 2017; 3276806
Jiang W, Zhang Y, Kane KT, et al. CD44 regulates pancreatic cancer invasion through MT1-MMP. Mol Cancer Res. 2015;13:9.
Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460.
Shi X, Guo X, Li X, et al. Loss of Linc01060 induces pancreatic cancer progression through vinculin-mediated focal adhesion turnover. Cancer Lett. 2018;31:10–20.
Reichert M, Takano S, Burstin JV, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013;27:288–300.
Chen NM, Singh G, Koenig A, et al. NFATc1 Links EGFR signaling to induction of Sox9 transcription and acinar-ductal transdifferentiation in the pancreas. Gastroenterology. 2015;148:1024–1034.
Lankisch PG, Breuer N, Bruns A, et al. Natural history of acute pancreatitis: a long-term population-based study. Am J Gastroenterol. 2009;104:2797–2805.
Pang Y, Kartsonaki C, Turnbull I, et al. Metabolic and lifestyle risk factors for acute pancreatitis in Chinese adults: a prospective cohort study of 0.5 million people. PLoS Med. 2018;15:e1002618.
KirkegÅRd J, Cronin-Fenton D, Heide-JØRgensen U, et al. Acute pancreatitis and pancreatic cancer risk: a nationwide matched-cohort study in denmark. Gastroenterology. 2018;154:1729.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fan, L., Hui, X., Mao, Y. et al. Identification of Acute Pancreatitis-Related Genes and Pathways by Integrated Bioinformatics Analysis. Dig Dis Sci 65, 1720–1732 (2020). https://doi.org/10.1007/s10620-019-05928-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-019-05928-5