Abstract
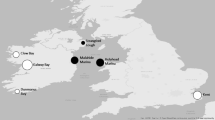
The decline of the eastern oyster (Crassostrea virginica) has prompted various restoration and aquaculture efforts. Recent field surveys in Rhode Island suggest that wild populations are increasing, yet the factors contributing to expansion are unknown. We used a population genetic approach to characterize genetic differences between wild and cultured oyster populations and explore the extent of connectivity and admixture between groups. Individual oysters from four wild, three farmed, and two restored populations were collected within or just outside Ninigret Pond, a coastal lagoon highly influenced by human activity, and genotyped at 13 microsatellite loci. Results from the multi-locus genotype data showed that wild populations were more genetically diverse than the cultured populations. We also observed significant genetic differentiation between paired wild and cultured populations but not between pairs of wild populations. A cluster analysis detected substantial admixture between wild and cultured groups. As oyster aquaculture and restoration activities are forecasted to increase in the future, this study highlights the potential degree of genetic introgression between remnant wild populations and less diverse, hatchery-reared stocks. Those tasked with preserving our living natural resources should carefully consider how the juxtaposition of aquaculture, restored, and wild populations at small spatial scales will impact the genetic composition and evolutionary trajectories of species in decline for generations to come.






Similar content being viewed by others
References
Anderson JD, Karel WJ, Mace CE, Bartram BL, Hare MP (2014) Spatial genetic features of eastern oyster (Cassostrea virginica Gmelin) in the Gulf of Mexico: northward movement of a secondary contact zone. Ecol Evol 4:1671–1683
Araki H, Schmid C (2010) Is hatchery stocking a help or harm? Aquaculture 308:S2–S11
Aranishi F, Okimoto T (2006) A simple and reliable method for DNA extraction from bivalve mantle. J Appl Genet 47:251–254
Arnaud-Haond S, Vonau V, Bonhomme F et al (2004) Spatio-temporal variation in the genetic composition of wild populations of pearl oyster (Pinctada margaritifera cumingii) in French Polynesia following 10 years of juvenile translocation. Mol Ecol 13:2001–2007
Beck MW, Brumbaugh RD, Airoldi L et al (2011) Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61:107–116
Buetel D (2014) Aquaculture in Rhode Island 2014 annual status report. http://www.crmc.ri.gov/aquaculture/aquareport14.pdf. Accessed 19 Dec 2017
Camara MD, Vadopalas B (2009) Genetic aspects of restoring olympia oysters and other native bivalves: balancing the need for action, good intentions, and the risks of making things worse. J Shellfish Res 28:121–145
Carlsson J (2008) Effects of microsatellite null alleles on assignment testing. J Hered 99:616–623
Carlsson J, Morrison CL, Reece KS (2006) Wild and aquaculture populations of the eastern oyster compared using microsatellites. J Hered 97:595–598
Carlsson J, Carnegie RB, Cordes JF et al (2008) Evaluating recruitment contribution of a selectively bred aquaculture line of the oyster, Crassostrea virginica used in restoration efforts. J Shellfish Res 27:1117–1124
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Coen LD, Humphries AT (2017) Oyster-generated marine habitats: their services, enhancement, restoration and monitoring. In: Allison SK, Murphy SD (eds) The Routledge handbook of ecological and environmental restoration. Routledge, New York, pp 274–294
Conover DO (1998) Local adaptation in marine fishes: evidence and implications for stock enhancement. Bull Mar Sci 62:477–493
Cooper AM, Miller LM, Kapuscinski AR (2009) Conservation of population structure and genetic diversity under captive breeding of remnant coaster brook trout (Salvelinus fontinalis) populations. Conserv Genet 11:1087–1093
Do C, Waples RS, Peel D et al (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Duball CE (2017) Environmental impacts of oyster aquaculture on the coastal lagoons of southern Rhode Island. Master Thesis, University of Rhode Island
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Falk DA, Richards CM, Zedler JB (2006) Integrating restoration ecology and ecological theory: a synthesis. In: Falk DA, Palmer MA, Zedler JB (eds) Foundations of restoration ecology. Island Press, Washington, DC, pp 341–346
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, red List criteria and population viability analyses. Biol Conserv 170:56–63
Gaffney PM (2006) The role of genetics in shellfish restoration. Aquat Living Resour 19:277–282
Grabowski JH, Peterson CH (2007) Restoring oyster reefs to recover ecosystem services. In: Cuddington K, Byers JE, Wilson WG, Hastings A (eds) Theoretical ecology series. Academic Press, Burlington, pp 281–298
Grant WS, Stewart Grant W, Jasper J et al (2017) Responsible genetic approach to stock restoration, sea ranching and stock enhancement of marine fishes and invertebrates. Rev Fish Biol Fish 27:615–649
Griffin M (2016) Fifteen years of Rhode Island oyster restoration: a performance evaluation and cost-benefit analysis. Master Thesis, University of Rhode Island
Gruenthal KM, Witting DA, Ford T et al (2013) Development and application of genomic tools to the restoration of green abalone in southern California. Conserv Genet 15:109–121
Guo X (2009) Use and exchange of genetic resources in molluscan aquaculture. Rev Aquac 1:251–259
Hanley TC, Hughes AR, Williams B et al (2016) Effects of intraspecific diversity on survivorship, growth, and recruitment of the eastern oyster across sites. Ecology 97:1518–1529
Hare MP, Allen SK Jr, Bloomer P, Camara MD, Carnegie RB, Murfree J, Luckenbach M, Meritt D, Morrison C, Paynter K, Reece KS, Rose CG (2006) A genetic test for recruitment enhancement in Chesapeake Bay oysters, Crassostrea virginica, after population supplementation with a disease tolerant strain. Conserv Genet 7:717–734
Hare MP, Nunney L, Schwartz MK et al (2011) Understanding and estimating effective population size for practical application in marine species management. Conserv Biol 25:438–449
He Y, Ford SE, Bushek D et al (2012) Effective population sizes of eastern oyster Crassostrea virginica (Gmelin) populations in Delaware Bay, USA. J Mar Res 70:357–379
Hedgecock D (1994) Does variance in reproductive success limit effective population sizes of marine organisms? In: Beaumont AR (ed) Genetics and evolution of aquatic organisms. Chapman and Hall, London, pp 122–134
Hoffman JI, Amos W (2005) Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol Ecol 14:599–612
Hughes AR, Stachowicz JJ (2004) Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc Natl Acad Sci USA 101:8998–9002
Jackson JB, Kirby MX, Berger WH et al (2001) Historical overfishing and the recent collapse of coastal ecosystems. Science 293:629–637
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jorde PE, Ryman N (2007) Unbiased estimator for genetic drift and effective population size. Genetics 177:927–935
Kenchington EL, Patwary MU, Zouros E, Bird CJ (2006) Genetic differentiation in relation to marine landscape in a broadcast-spawning bivalve mollusc (Placopecten magellanicus). Mol Ecol 15:1781–1796
Kirby MX (2004) Fishing down the coast: historical expansion and collapse of oyster fisheries along continental margins. Proc Natl Acad Sci USA 101:13096–13099
Kitada S (2018) Economic, ecological, and genetic impacts of marine stock enhancement and sea ranching: a systematic review. Fish Fish 19:511–532
Kochmann J, Carlsson J, Crowe TP, Mariani S (2012) Genetic evidence for the uncoupling of local aquaculture activities and a population of an invasive species—a case study of pacific oysters (Crassostrea gigas). J Hered 103:661–671
Levin LA (2006) Recent progress in understanding larval dispersal: new directions and digressions. Integr Comp Biol 46:282–297
Lind CE, Evans BS, Knauer J et al (2009) Decreased genetic diversity and a reduced effective population size in cultured silver-lipped pearl oysters (Pinctada maxima). Aquaculture 286:12–19
Matschiner M, Salzburger W (2009) TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25:1982–1983
Milbury CA, Meritt DW, Newell RIE, Gaffney PM (2004) Mitochondrial DNA markers allow monitoring of oyster stock enhancement in the Chesapeake Bay. Mar Biol. https://doi.org/10.1007/s00227-004-1312-z
Naylor RL, Goldburg RJ, Primavera JH et al (2000) Effect of aquaculture on world fish supplies. Nature 405:1017–1024
Nixon SW, Buckley B, Granger S, Bintz J (2001) Responses of very shallow marine ecosystems to nutrient enrichment. Hum Ecol Assess 7:1457–1481
Palumbi SR (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13:146–158
Paquette SR (2011) PopGenKit: useful functions for (batch) file conversion and data resampling in microsatellite data sets. R package version 1.0. http://CRAN.R-project.org/package=PopGenKit. Accessed 22 Oct 2017
Peters JW, Eggleston DB, Puckett BJ, Theuerkauf SJ (2017) Oyster demographics in harvested reefs vs. no-take reserves: Implications for larval spillover and restoration success. Front Mar Sci. https://doi.org/10.3389/fmars.2017.00326
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proestou DA, Vinyard BT, Corbett RJ et al (2016) Performance of selectively-bred lines of eastern oyster, Crassostrea virginica, across eastern US estuaries. Aquaculture 464:17–27
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reece KS, Ribeiro WL, Gaffney PM et al (2004) Microsatellite marker development and analysis in the eastern oyster (Crassostrea virginica): confirmation of null alleles and non-Mendelian segregation ratios. J Hered 95:346–352
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Rice MA, Valliere A, Caporelli A (2000) A review of shellfish restoration and management projects in Rhode Island. J Shellfish Res 19:401–408
Rose CG, Paynter KT, Hare MP (2006) Isolation by distance in the eastern oyster, Crassostrea virginica, in Chesapeake Bay. J Hered 97:158–170
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Sanford E, Kelly MW (2011) Local adaptation in marine invertebrates. Annu Rev Mar Sci 3:509–535
Schindler DE, Hilborn R, Chasco B et al (2010) Population diversity and the portfolio effect in an exploited species. Nature 465:609–612
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Smee DL, Overath RD, Johnson KD, Sanchez JA (2013) Intraspecific variation influences natural settlement of eastern oysters. Oecologia 173:947–953
Spires JE (2015) The exchange of eastern oyster (Crassostrea virginica) larvae between subpopulations in the Choptank and Little Choptank rivers: model simulations, the influence of salinity, and implications for restoration. Master Thesis, University of Maryland
Thompson JA, Stow AJ, Raftos DA (2017) Lack of genetic introgression between wild and selectively bred Sydney rock oysters Saccostrea glomerata. Mar Ecol Prog Ser 570:127–139
Thorpe JP, Solé-Cava AM, Watts PC (2000) Exploited marine invertebrates: genetics and fisheries. In: Solé-Cava AM, Russo CAM, Thorpe JP (eds) Marine genetics. Springer, Dordrecht, pp 165–184
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang Y, Guo X (2007) Development and characterization of EST-SSR markers in the eastern oyster Crassostrea virginica. Mar Biotechnol 9:500–511
Wang Y, Wang X, Wang A, Guo X (2010) A 16-microsatellite multiplex assay for parentage assignment in the eastern oyster (Crassostrea virginica Gmelin). Aquaculture 308:S28–S33
Waples RS (1989) A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121:379–391
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184
Waples RS (2016) Tiny estimates of the Ne/N ration in marine fishes: are they real? J Fish Biol 89:2479–2504
Waples RS (2018) Null alleles and FIS x FST correlations. J Hered 109:457–461
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262
Ward RD (2006) The importance of identifying spatial population structure in restocking and stock enhancement programmes. Fish Res 80:9–18
Weersing K, Toonen RJ (2009) Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser 393:1–12
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Zu Ermgassen PSE, Zu PS, Spalding MD et al (2012) Historical ecology with real numbers: past and present extent and biomass of an imperilled estuarine habitat. Proc R Soc B 279:3393–3400
Acknowledgements
We thank Dr. Marta Gómez-Chiarri for access to laboratory space and equipment, and Jessica Piez and Saebom Sohn for assistance in the lab. Dillon McNulty, Bray Beltran, and Jeanne Parente helped with oyster collections in the field. We also thank Mary Sullivan for help with GIS, Eric Schneider and Gary Casabona for useful discussions about shellfish restoration in RI, and two anonymous reviewers for very thoughtful and constructive comments on this manuscript. This work was funded through USDA ARS CRIS Project #803031000003, The Nature Conservancy’s GLOBE internship program, and a non-competitive grant from the USDA NRCS. Access to the University of Rhode Island Genomics and Sequencing Center, which is supported in part by the National Science Foundation under EPSCoR Cooperative Agreement # EPS-1004057, was also instrumental for the completion of this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jaris, H., Brown, D.S. & Proestou, D.A. Assessing the contribution of aquaculture and restoration to wild oyster populations in a Rhode Island coastal lagoon. Conserv Genet 20, 503–516 (2019). https://doi.org/10.1007/s10592-019-01153-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01153-9