Abstract
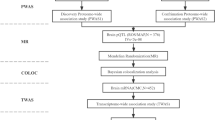
Amyotrophic lateral sclerosis, a fatal neurodegeneration disease affecting motor neurons in the brain and spinal cord, is difficult to diagnose and treat. The objective of this study is to identify novel candidate genes related to ALS. Transcriptome-wide association study of ALS was conducted by integrating the genome-wide association study summary data (including 1234 ALS patients and 2850 controls) and pre-computed gene expression weights of different tissues. The ALS-associated genes identified by TWAS were further compared with the differentially expressed genes detected by the mRNA expression profiles of the sporadic ALS. Functional enrichment and annotation analysis of identified genes were performed by an R package and the functional mapping and annotation software. TWAS identified 761 significant genes (PTWAS < 0.05), 627 Gene ontology terms, and 8 Kyoto Encyclopedia of Genes and Genomes pathways for ALS, such as C9orf72, with three expression quantitative trait loci were found significantly: rs2453554 (PTWAS CBRS = 4.68 × 10–10, PTWAS CBRS = 2.54 × 10–9), rs10967976 (PTWAS CBRS = 7.85 × 10–10, PTWAS CBRS = 8.91 × 10–9, PTWAS CBRS = 1.49 × 10–7, PTWAS CBRS = 5.59 × 10–7), rs3849946 (PTWAS CBRS = 7.69 × 10–4, PTWAS YBL = 4.02 × 10–2), Mitochondrion (Padj = 4.22 × 10–16), and Cell cycle (Padj = 2.04 × 10–3). Moreover, 107 common genes, 4 KEGG pathways and 41 GO terms were detected by integrating mRNA expression profiles of sALS, such as CPVL (FC = 2.06, PmRNA = 6.99 × 10–6, PTWAS CBR = 2.88 × 10–2, PTWAS CBR = 4.37 × 10–2), Pyrimidine Metabolism (Padj = 2.43 × 10–2), and Cell Activation (Padj = 5.54 × 10–3). Multiple candidate genes and pathways were detected for ALS. Our findings may provide novel clues for understanding the genetic mechanism of ALS.




Similar content being viewed by others
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 121(Pt 22):3778–3785. https://doi.org/10.1242/jcs.038950
Balendra R, Isaacs AM (2018) C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol 14(9):544–558. https://doi.org/10.1038/s41582-018-0047-2
Benyamin B, He J, Zhao Q, Gratten J, Garton F, Leo PJ, Liu Z, Mangelsdorf M, Al-Chalabi A, Anderson L, Butler TJ, Chen L, Chen XD, Cremin K, Deng HW, Devine M, Edson J, Fifita JA, Furlong S, Han YY, Harris J, Henders AK, Jeffree RL, Jin ZB, Li Z, Li T, Li M, Lin Y, Liu X, Marshall M, McCann EP, Mowry BJ, Ngo ST, Pamphlett R, Ran S, Reutens DC, Rowe DB, Sachdev P, Shah S, Song S, Tan LJ, Tang L, van den Berg LH, van Rheenen W, Veldink JH, Wallace RH, Wheeler L, Williams KL, Wu J, Wu X, Yang J, Yue W, Zhang ZH, Zhang D, Noakes PG, Blair IP, Henderson RD, McCombe PA, Visscher PM, Xu H, Bartlett PF, Brown MA, Wray NR, Fan D (2017) Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun 8(1):611. https://doi.org/10.1038/s41467-017-00471-1
Brown RH, Al-Chalabi A (2017) Amyotrophic lateral sclerosis. N Engl J Med 377(2):162–172. https://doi.org/10.1056/NEJMra1603471
D’Ovidio F, d’Errico A, Carnà P, Calvo A, Costa G, Chiò A (2018) The role of pre-morbid diabetes on developing amyotrophic lateral sclerosis. Eur J Neurol 25(1):164–170. https://doi.org/10.1111/ene.13465
Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, de Jong B, Rigouts L, Sloutsky A, Kaur D, Sunyaev S, van Soolingen D, Shendure J, Sacchettini J, Murray M (2019) GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun 10(1):2128. https://doi.org/10.1038/s41467-019-10110-6
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimäki T, Raitoharju E, Kähönen M, Seppälä I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48(3):245–252. https://doi.org/10.1038/ng.3506
Hartley JN, Simard LR, Ly V, Del Bigio MR, Frosk P (2019) A homozygous canonical splice acceptor site mutation in PRUNE1 is responsible for a rare childhood neurodegenerative disease in Manitoba Cree families. Am J Med Genet A 179(2):206–218. https://doi.org/10.1002/ajmg.a.60690
Huang H, Song Y, Wu Y, Guo N, Ma Y, Qian L (2015) Erbin loss promotes cancer cell proliferation through feedback activation of Akt-Skp2-p27 signaling. Biochem Biophys Res Commun 463(3):370–376. https://doi.org/10.1016/j.bbrc.2015.05.071
Imagawa E, Yamamoto Y, Mitsuhashi S, Isidor B, Fukuyama T, Kato M, Sasaki M, Tanabe S, Miyatake S, Mizuguchi T, Takata A, Miyake N, Matsumoto N (2018) PRUNE1-related disorder: expanding the clinical spectrum. Clin Genet 94(3–4):362–367. https://doi.org/10.1111/cge.13385
Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, Emokpae R Jr, Gorospe M, Mattson MP (2004) Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41(4):549–561. https://doi.org/10.1016/s0896-6273(04)00017-0
Li P, Ning Y, Guo X, Wen Y, Cheng B, Ma M, Zhang L, Cheng S, Wang S, Zhang F (2019) Integrating transcriptome-wide study and mRNA expression profiles yields novel insights into the biological mechanism of chondropathies. Arthritis Res Ther 21(1):194. https://doi.org/10.1186/s13075-019-1978-8
Liang C, Tao Y, Shen C, Tan Z, Xiong WC, Mei L (2012) Erbin is required for myelination in regenerated axons after injury. J Neurosc 32(43):15169–15180. https://doi.org/10.1523/jneurosci.2466-12.2012
Marin B, Boumédiene F, Logroscino G, Couratier P, Babron MC, Leutenegger AL, Copetti M, Preux PM, Beghi E (2017) Variation in worldwide incidence of amyotrophic lateral sclerosis: a meta-analysis. Int J Epidemiol 46(1):57–74. https://doi.org/10.1093/ije/dyw061
Mariosa D, Kamel F, Bellocco R, Ye W, Fang F (2015) Association between diabetes and amyotrophic lateral sclerosis in Sweden. Eur J Neurol 22(11):1436–1442. https://doi.org/10.1111/ene.12632
Masrori P, Van Damme P (2020) Amyotrophic lateral sclerosis: a clinical review. Eur J Neurol 27(10):1918–1929. https://doi.org/10.1111/ene.14393
McCauley ME, O’Rourke JG, Yáñez A, Markman JL, Ho R, Wang X, Chen S, Lall D, Jin M, Muhammad A, Bell S, Landeros J, Valencia V, Harms M, Arditi M, Jefferies C, Baloh RH (2020) C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature 585(7823):96–101. https://doi.org/10.1038/s41586-020-2625-x
Neale JH, Olszewski RT, Gehl LM, Wroblewska B, Bzdega T (2005) The neurotransmitter N-acetylaspartylglutamate in models of pain, ALS, diabetic neuropathy, CNS injury and schizophrenia. Trends Pharmacol Sci 26(9):477–484. https://doi.org/10.1016/j.tips.2005.07.004
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314(5796):130–133. https://doi.org/10.1126/science.1134108
Oskarsson B, Gendron TF, Staff NP (2018) Amyotrophic lateral sclerosis: an update for 2018. Mayo Clin Proc 93(11):1617–1628. https://doi.org/10.1016/j.mayocp.2018.04.007
Peng S, Deyssenroth MA, Di Narzo AF, Lambertini L, Marsit CJ, Chen J, Hao K (2017) Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum Mol Genet 26(17):3432–3441. https://doi.org/10.1093/hmg/ddx265
Qi X, Yu F, Wen Y, Li P, Cheng B, Ma M, Cheng S, Zhang L, Liang C, Liu L, Zhang F (2020) Integration of transcriptome-wide association study and messenger RNA expression profile to identify genes associated with osteoarthritis. Bone Joint Res 9(3):130–138. https://doi.org/10.1302/2046-3758.93.Bjr-2019-0137.R1
Rabin SJ, Kim JM, Baughn M, Libby RT, Kim YJ, Fan Y, Libby RT, La Spada A, Stone B, Ravits J (2010) Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet 19(2):313–328. https://doi.org/10.1093/hmg/ddp498
Schwartz EI, Smilenov LB, Price MA, Osredkar T, Baker RA, Ghosh S, Shi FD, Vollmer TL, Lencinas A, Stearns DM, Gorospe M, Kruman II (2007) Cell cycle activation in postmitotic neurons is essential for DNA repair. Cell Cycle (georgetown, Tex) 6(3):318–329. https://doi.org/10.4161/cc.6.3.3752
Singh A, Kukreti R, Saso L, Kukreti S (2019) Oxidative stress: a key modulator in neurodegenerative diseases. Molecules. https://doi.org/10.3390/molecules24081583
van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, van der Spek RA, Võsa U, de Jong S, Robinson MR, Yang J, Fogh I, van Doormaal PT, Tazelaar GH, Koppers M, Blokhuis AM, Sproviero W, Jones AR, Kenna KP, van Eijk KR, Harschnitz O, Schellevis RD, Brands WJ, Medic J, Menelaou A, Vajda A, Ticozzi N, Lin K, Rogelj B, Vrabec K, Ravnik-Glavač M, Koritnik B, Zidar J, Leonardis L, Grošelj LD, Millecamps S, Salachas F, Meininger V, de Carvalho M, Pinto S, Mora JS, Rojas-García R, Polak M, Chandran S, Colville S, Swingler R, Morrison KE, Shaw PJ, Hardy J, Orrell RW, Pittman A, Sidle K, Fratta P, Malaspina A, Topp S, Petri S, Abdulla S, Drepper C, Sendtner M, Meyer T, Ophoff RA, Staats KA, Wiedau-Pazos M, Lomen-Hoerth C, Van Deerlin VM, Trojanowski JQ, Elman L, McCluskey L, Basak AN, Tunca C, Hamzeiy H, Parman Y, Meitinger T, Lichtner P, Radivojkov-Blagojevic M, Andres CR, Maurel C, Bensimon G, Landwehrmeyer B, Brice A, Payan CA, Saker-Delye S, Dürr A, Wood NW, Tittmann L, Lieb W, Franke A, Rietschel M, Cichon S, Nöthen MM, Amouyel P, Tzourio C, Dartigues JF, Uitterlinden AG, Rivadeneira F, Estrada K, Hofman A, Curtis C, Blauw HM, van der Kooi AJ, de Visser M, Goris A, Weber M, Shaw CE, Smith BN, Pansarasa O, Cereda C, Del Bo R, Comi GP, D’Alfonso S, Bertolin C, Sorarù G, Mazzini L, Pensato V, Gellera C, Tiloca C, Ratti A, Calvo A, Moglia C, Brunetti M, Arcuti S, Capozzo R, Zecca C, Lunetta C, Penco S, Riva N, Padovani A, Filosto M, Muller B, Stuit RJ, Blair I, Zhang K, McCann EP, Fifita JA, Nicholson GA, Rowe DB, Pamphlett R, Kiernan MC, Grosskreutz J, Witte OW, Ringer T, Prell T, Stubendorff B, Kurth I, Hübner CA, Leigh PN, Casale F, Chio A, Beghi E, Pupillo E, Tortelli R, Logroscino G, Powell J, Ludolph AC, Weishaupt JH, Robberecht W, Van Damme P, Franke L, Pers TH, Brown RH, Glass JD, Landers JE, Hardiman O, Andersen PM, Corcia P, Vourc’h P, Silani V, Wray NR, Visscher PM, de Bakker PI, van Es MA, Pasterkamp RJ, Lewis CM, Breen G, Al-Chalabi A, van den Berg LH, Veldink JH (2016) Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 48(9):1043–1048. https://doi.org/10.1038/ng.3622
Vazquez-Villaseñor I, Garwood CJ, Heath PR, Simpson JE, Ince PG, Wharton SB (2020) Expression of p16 and p21 in the frontal association cortex of ALS/MND brains suggests neuronal cell cycle dysregulation and astrocyte senescence in early stages of the disease. Neuropathol Appl Neurobiol 46(2):171–185. https://doi.org/10.1111/nan.12559
Vincenzetti S, Polzonetti V, Micozzi D, Pucciarelli S (2016) Enzymology of pyrimidine metabolism and neurodegeneration. Curr Med Chem 23(14):1408–1431. https://doi.org/10.2174/0929867323666160411125803
Wallings RL, Humble SW, Ward ME, Wade-Martins R (2019) Lysosomal dysfunction at the centre of parkinson’s disease and frontotemporal dementia/amyotrophic lateral sclerosis. Trends Neurosci 42(12):899–912. https://doi.org/10.1016/j.tins.2019.10.002
Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8(1):1826. https://doi.org/10.1038/s41467-017-01261-5
Xiao L, Yuan Z, Jin S, Wang T, Huang S, Zeng P (2020) Multiple-tissue integrative transcriptome-wide association studies discovered new genes associated with amyotrophic lateral sclerosis. Front Genet 11:587243. https://doi.org/10.3389/fgene.2020.587243
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Zhang QJ, Chen Y, Zou XH, Hu W, Ye ML, Guo QF, Lin XL, Feng SY, Wang N (2020) Promoting identification of amyotrophic lateral sclerosis based on label-free plasma spectroscopy. Ann Clin Trans Neurol 7(10):2010–2018. https://doi.org/10.1002/acn3.51194
Zou ZY, Zhou ZR, Che CH, Liu CY, He RL, Huang HP (2017) Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 88(7):540–549. https://doi.org/10.1136/jnnp-2016-315018
Acknowledgements
This work was supported by the National Natural Science Foundation of China [81922059]; and the Fundamental Research Funds for the Central Universities [xzy022019006]. We would like to thank the authors who generously shared their data as well as all study participants.
Author information
Authors and Affiliations
Contributions
FZ and ZMZ take responsibility for the integrity of the work as a whole, from inception to the finished manuscript. ZMZ gave the conception, design and some critical suggestion to this study. PL, SQC, BLC, and LL performed all analysis. XHW, XA, ZCH, CRL, and SEL partially performed some experiments. PL and SQC convinced the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
Not applicable.
Consent for Publication
All authors consent to the publication of current data.
Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, P., Cheng, S., Wen, Y. et al. Identifying Candidate Genes Associated with Sporadic Amyotrophic Lateral Sclerosis via Integrative Analysis of Transcriptome-Wide Association Study and Messenger RNA Expression Profile. Cell Mol Neurobiol 43, 327–338 (2023). https://doi.org/10.1007/s10571-021-01186-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-021-01186-0