Abstract
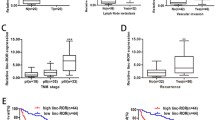
Adriamycin is widely used as a chemotherapeutic strategy for advanced hepatocellular carcinoma (HCC). However, the clinical response was disappointing because of the acquired drug resistance with long-term usage. Revealing the underlying mechanism could provide promising therapeutics for the drug-resistant patients. The recently identified linc-ROR (long intergenic non-protein-coding RNA, regulator of reprogramming) has been found to be an oncogene in various cancers, and it also demonstrated to mediate drug resistance and metastasis. We thereby wonder whether this lincRNA could mediate adriamycin chemoresistance in HCC. In this study, linc-ROR was found to be upregulated in adriamycin-resistant HCC cells. And its overexpression accelerated epithelial-mesenchymal transition (EMT) program and adriamycin resistance. Conversely, its silence suppressed EMT and made HCC cells sensitize to adriamycin in vitro and in vivo. Further investigation revealed that linc-ROR physically interacted with AP-2α, mediated its stability by a post-translational modification manner, and sequentially activated Wnt/β-catenin pathway. Furthermore, linc-ROR expression was positively associated with β-catenin expression in human clinical specimens. Taken together, linc-ROR promoted tumorigenesis and adriamycin resistance in HCC via a linc-ROR/AP-2α/Wnt/β-catenin axis, which could be developed as a potential therapeutic target for the adriamycin-resistant patients.
Graphical Abstract









Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AP-2α:
-
Activator protein 2α
- CRC:
-
Colorectal cancer
- c-myc:
-
MYC proto-oncogene
- CCND1:
-
Cyclin D1
- CD44:
-
CD44 molecule
- CHX:
-
Cycloheximide
- CCK8:
-
Cell Counting Assay Kit-8
- CSC:
-
Cancer stem cell
- CCAL:
-
Colorectal cancer-associated lncRNA
- EMT:
-
Epithelial-mesenchymal transition
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- HCC:
-
Hepatocellular carcinoma
- HULC:
-
LncRNA highly upregulated in liver cancer
- HOTTIP:
-
The lncRNA HOXA transcript at the distal tip
- iPSCs:
-
Induced pluripotent stem cells
- lncRNA:
-
Long non-coding RNA
- lncFZD6:
-
The lncRNA frizzled 6
- MDR:
-
Multidrug resistance
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- NEAT1:
-
Nuclear-enriched abundant transcript 1
- NC:
-
Negative control
- Oct3/4:
-
Organic cation/carnitine transporter 3
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RIP:
-
RNA immunoprecipitation
- ROR:
-
Regulator of reprogramming
- SD:
-
Standard deviation
- TACE:
-
Transarterial chemoembolization
- VEGF:
-
Vascular endothelial growth factor
References
Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19(1):39–56. https://doi.org/10.1038/s41573-019-0044-1.
Chen W, Yang J, Fang H, Li L, Sun J. Relevance function of Linc-ROR in the pathogenesis of cancer. Front Cell Dev Biol. 2020;8:696. https://doi.org/10.3389/fcell.2020.00696.
Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–205. https://doi.org/10.1016/j.cell.2012.05.012.
Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21(7):965. https://doi.org/10.3390/molecules21070965.
Eckert D, Buhl S, Weber S, Jäger R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6(13):246. https://doi.org/10.1186/gb-2005-6-13-246.
Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Updat. 2020;53: 100715. https://doi.org/10.1016/j.drup.2020.100715.
Feng L, Shi L, Lu YF, Wang B, Tang T, Fu WM, … Zhang JF. Linc-ROR promotes osteogenic differentiation of mesenchymal stem cells by functioning as a competing endogenous RNA for miR-138 and miR-145. Mol Ther Nucleic Acids. 2018;11:345–53. https://doi.org/10.1016/j.omtn.2018.03.004.
Forner A, Gilabert M, Bruix J, Raoul JL. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol. 2014;11(9):525–35. https://doi.org/10.1038/nrclinonc.2014.122.
Gao Y, Chen X, Zhang J. LncRNA MEG3 inhibits retinoblastoma invasion and metastasis by inducing β-catenin degradation. Am J Cancer Res. 2022;12(7):3111–27.
Ge L, Vujanovic NL. Soluble TNF regulates TACE via AP-2α transcription factor in mouse dendritic cells. J Immunol. 2017;198(1):417–27. https://doi.org/10.4049/jimmunol.1600524.
He Y, Jiang X, Duan L, Xiong Q, Yuan Y, Liu P, … Chen Y. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer. 2021;20(1):156. https://doi.org/10.1186/s12943-021-01469-6.
Huang CX, Jiang ZX, Du DY, Zhang ZM, Liu Y, Li YT. Hsa_circ_0016070/micro-340-5p axis accelerates pulmonary arterial hypertension progression by upregulating TWIST1 transcription via TCF4/β-catenin complex. J Am Heart Assoc. 2022;11(14): e024147. https://doi.org/10.1161/jaha.121.024147.
Huang W, Chen C, Liang Z, Qiu J, Li X, Hu X, … Zhang J. AP-2α inhibits hepatocellular carcinoma cell growth and migration. Int J Oncol. 2016;48(3):1125–34. https://doi.org/10.3892/ijo.2016.3318.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. https://doi.org/10.1038/nm.3981.
Kołat D, Kałuzińska Ż, Bednarek AK, Płuciennik E. The biological characteristics of transcription factors AP-2α and AP-2γ and their importance in various types of cancers. Biosci Rep. 2019;39(3):BSR20181928. https://doi.org/10.1042/BSR20181928.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. https://doi.org/10.1016/j.cell.2018.01.011.
Li M, Wang Y, Hung MC, Kannan P. Inefficient proteasomal-degradation pathway stabilizes AP-2alpha and activates HER-2/neu gene in breast cancer. Int J Cancer. 2006;118(4):802–11. https://doi.org/10.1002/ijc.21426.
Li Q, Dashwood RH. Activator protein 2alpha associates with adenomatous polyposis coli/beta-catenin and Inhibits beta-catenin/T-cell factor transcriptional activity in colorectal cancer cells. J Biol Chem. 2004;279(44):45669–75. https://doi.org/10.1074/jbc.M405025200.
Lian W, Zhang L, Yang L, Chen W. AP-2α reverses vincristine-induced multidrug resistance of SGC7901 gastric cancer cells by inhibiting the Notch pathway. Apoptosis. 2017;22(7):933–41. https://doi.org/10.1007/s10495-017-1379-x.
Liang W, Shi C, Hong W, Li P, Zhou X, Fu W, … Zhang J. Super-enhancer-driven lncRNA-DAW promotes liver cancer cell proliferation through activation of Wnt/β-catenin pathway. Mol Ther Nucleic Acids. 2021;26:1351–63. https://doi.org/10.1016/j.omtn.2021.10.028.
Llovet J, Kelley R, Villanueva A, Singal A, Pikarsky E, Roayaie S, … Finn RJNRDP. Hepatocell Carcinoma 2021;7(1):6. doi:https://doi.org/10.1038/s41572-020-00240-3
Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, … Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–7. https://doi.org/10.1038/ng.710.
Lou Y, Jiang H, Cui Z, Wang L, Wang X, Tian T. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/β-catenin signaling. Oncotarget. 2017;8(41):69983–94. https://doi.org/10.18632/oncotarget.19545.
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, … Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65(9):1494–504. https://doi.org/10.1136/gutjnl-2014-308392.
Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–9. https://doi.org/10.3322/caac.21161.
Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23(21):4776–89. https://doi.org/10.1200/jco.2005.05.081.
Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–99. https://doi.org/10.1016/j.cell.2017.05.016.
Pan Y, Chen J, Tao L, Zhang K, Wang R, Chu X, Chen L. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2017;8(20):33144–58. https://doi.org/10.18632/oncotarget.16562.
Peng WX, Huang JG, Yang L, Gong AH, Mo YY. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol Cancer. 2017;16(1):161. https://doi.org/10.1186/s12943-017-0727-3.
Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, … Banales JM. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121–36. https://doi.org/10.1038/s41575-018-0075-9.
Qiao MX, Li C, Zhang AQ, Hou LL, Yang J, Hu HG. Regulation of DEK expression by AP-2α and methylation level of DEK promoter in hepatocellular carcinoma. Oncol Rep. 2016;36(4):2382–90. https://doi.org/10.3892/or.2016.4984.
Ren P, Sheng Z, Wang Y, Yi X, Zhou Q, Zhou J, … Zhang J. RNF20 promotes the polyubiquitination and proteasome-dependent degradation of AP-2α protein. Acta Biochim Biophys Sin (shanghai). 2014;46(2):136–40. https://doi.org/10.1093/abbs/gmt136.
Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–29. https://doi.org/10.1038/nrclinonc.2017.44.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. https://doi.org/10.1038/s41580-020-00315-9.
Sun Z, Sun D, Feng Y, Zhang B, Sun P, Zhou B, … Zhan H. Exosomal linc-ROR mediates crosstalk between cancer cells and adipocytes to promote tumor growth in pancreatic cancer. Mol Ther Nucleic Acids. 2021;26:253–68. https://doi.org/10.1016/j.omtn.2021.06.001.
Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–62. https://doi.org/10.1056/NEJMra1713263.
Wang L, Yu X, Zhang Z, Pang L, Xu J, Jiang J, … Li F. Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J Exp Clin Cancer Res. 2017;36(1):182. https://doi.org/10.1186/s13046-017-0658-2.
Wang M, Yu F, Chen X, Li P, Wang K. The underlying mechanisms of noncoding RNAs in the chemoresistance of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2020;21:13–27. https://doi.org/10.1016/j.omtn.2020.05.011.
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, … Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. https://doi.org/10.1016/j.devcel.2013.03.002.
Wei-Cheng L, Cheuk-Wa W, Pu-Ping L, Mai S, Ye C, Shi-Tao R, … Jin-Fang Z. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019. https://doi.org/10.1186/s13059-019-1685-4.
Xu T, Zhang J, Chen W, Pan S, Zhi X, Wen L, … Liang T. ARK5 promotes doxorubicin resistance in hepatocellular carcinoma via epithelial-mesenchymal transition. Cancer Lett. 2016;377(2):140–8. https://doi.org/10.1016/j.canlet.2016.04.026.
Yu N, Hu T, Yang H, Zhang L, Song Q, Xiang F, … Li Y. Twist1 contributes to the maintenance of some biological properties of dermal papilla cells in vitro by forming a complex with Tcf4 and β-catenin. Front Cell Dev Biol. 2020;8:824. https://doi.org/10.3389/fcell.2020.00824.
Zhang F, Chen K, Tao H, Kang T, Xiong Q, Zeng Q, … Chen M. miR-25–3p, positively regulated by transcription factor AP-2α, regulates the metabolism of C2C12 cells by targeting Akt1. Int J Mol Sci 2018;19(3). doi:https://doi.org/10.3390/ijms19030773
Zhang X, Chen J, Jiang S, He S, Bai Y, Zhu L, … Liang X. N-acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial-to-mesenchymal transition. Oxid Med Cell Longev. 2019;2019:7561879. https://doi.org/10.1155/2019/7561879.
Zhang Y, Wu W, Sun Q, Ye L, Zhou D, Wang W. linc-ROR facilitates hepatocellular carcinoma resistance to doxorubicin by regulating TWIST1-mediated epithelial-mesenchymal transition. Mol Med Rep. 2021;23(5):340. https://doi.org/10.3892/mmr.2021.11979.
Zhi Y, Abudoureyimu M, Zhou H, Wang T, Feng B, Wang R, Chu X. FOXM1-mediated LINC-ROR regulates the proliferation and sensitivity to sorafenib in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2019;16:576–88. https://doi.org/10.1016/j.omtn.2019.04.008.
Funding
This work was supported by the National Natural Science Foundation of China (81773066, 81772404) and Natural Science Foundation of Guangdong Province (2020A1515010961, 2021A1515012111).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Chuan-Jian Shi, Min-Yi Lv, Li-Qiang Deng, and Wei-Qiang Zeng. The experiments conceptualized and designed by Wei-Ming Fu and Jin-Fang Zhang. The first draft of the manuscript was written by Chuan-Jian Shi and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The usage and treatment of animals were approved by Institutional Animal Care and Use Committee (IACUC) of Southern Medical University (SMU, Guangzhou, China, Approval No. L2018187). All the tissues were obtained with informed consent and this study was approved by Joint Chinese University of Hong Kong-New Territories Ease Cluster Clinical Research Ethics Committee.
Consent to participate
Informed consent was obtained where applicable.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Linc-ROR and Wnt/β-catenin signaling were stimulated in an adriamycin-resistant HCC cells.
• Linc-ROR accelerated EMT programme and adriamycin resistance through activating the Wnt/β-catenin signaling in HCC cells.
• Linc-ROR physically interacted AP-2α, triggering its ubiquitin-mediated degradation and stimulating Wnt/β-catenin signaling in HCC cells.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, CJ., Lv, MY., Deng, LQ. et al. Linc-ROR drive adriamycin resistance by targeting AP-2α/Wnt/β-catenin axis in hepatocellular carcinoma. Cell Biol Toxicol 39, 1735–1752 (2023). https://doi.org/10.1007/s10565-022-09777-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-022-09777-3