Abstract
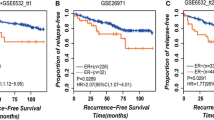
Current predictors for estrogen receptor-positive (ER-positive) breast cancer patients receiving tamoxifen are often invalid in inter-laboratory validation. We aim to develop a robust predictor based on the relative ordering of expression measurement (ROE) in gene pairs. Using a large integrated dataset of 420 normal controls and 1,129 ER-positive breast tumor samples, we identified the gene pairs with stable ROEs in normal control and significantly reversed ROEs in ER-positive tumor. Using these gene pairs, we characterized each sample of a cohort of 292 ER-positive patients who received tamoxifen monotherapy for 5 years and then identified relapse risk-associated gene pairs. We extracted a gene pair subset that resulted in the largest positive and negative predictive values for predicting 10-year relapse-free survival (RFS) using a genetic algorithm. A predictor was developed based on the gene pair subset and was validated in 2 large multi-laboratory cohorts (N = 250 and 248, respectively) of ER-positive patients who received 5-year tamoxifen alone. In the first validation cohort, the patients predicted to be tamoxifen sensitive had a 10-year RFS of 91 % (95 % confidence interval [CI] 85–97 %) with an absolute risk reduction of 34 % (95 % CI 17–51 %). The patients predicted to be tamoxifen insensitive had a significantly higher relapse risk than the patients predicted to be tamoxifen sensitive (hazard ratio = 4.99, 95 % CI 2.45–10.17, P = 9.13 × 10−7). Similar performance was achieved for the second validation cohort. The predictor performed well in both node-negative and node-positive subsets and added significant predictive power to the clinical parameters. In contrast, 2 previously proposed predictors did not achieve significantly better performances than the baselines of the validation cohorts. In summary, the proposed predictor can accurately and robustly predict tamoxifen sensitivity of ER-positive breast cancer patients and identified patients with a high probability of 10-year RFS following tamoxifen monotherapy.



Similar content being viewed by others
References
Hackshaw A, Roughton M, Forsyth S, Monson K, Reczko K, Sainsbury R, Baum M (2011) Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol 29(13):1657–1663. doi:10.1200/JCO.2010.32.2933
Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, Dowsett M, Ingle J, Peto R (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378(9793):771–784. doi:10.1016/S0140-6736(11)60993-8
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351(27):2817–2826. doi:10.1056/NEJMoa041588
Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5(6):607–616. doi:10.1016/j.ccr.2004.05.015
Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM (2006) Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24(11):1656–1664. doi:10.1200/JCO.2005.03.2755
Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, Gillet C, Ellis P, Ryder K, Reid JF, Daidone MG, Pierotti MA, Berns EM, Jansen MP, Foekens JA, Delorenzi M, Bontempi G, Piccart MJ, Sotiriou C (2008) Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genom 9:239. doi:10.1186/1471-2164-9-239
Symmans WF, Hatzis C, Sotiriou C, Andre F, Peintinger F, Regitnig P, Daxenbichler G, Desmedt C, Domont J, Marth C, Delaloge S, Bauernhofer T, Valero V, Booser DJ, Hortobagyi GN, Pusztai L (2010) Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol 28(27):4111–4119. doi:10.1200/JCO.2010.28.4273
Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM (2006) Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 355(6):560–569. doi:10.1056/NEJMoa052933
Iwamoto T, Pusztai L (2010) Predicting prognosis of breast cancer with gene signatures: are we lost in a sea of data? Genom Med 2(11):81. doi:10.1186/gm202
Buyse M, Sargent DJ, Grothey A, Matheson A, de Gramont A (2010) Biomarkers and surrogate end points—the challenge of statistical validation. Nat Rev Clin Oncol 7(6):309–317. doi:10.1038/nrclinonc.2010.43
Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS (2012) Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol 9(1):58–64. doi:10.1038/nrclinonc.2011.125
Prat A, Parker JS, Fan C, Cheang MC, Miller LD, Bergh J, Chia SK, Bernard PS, Nielsen TO, Ellis MJ, Carey LA, Perou CM (2012) Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann Oncol 23(11):2866–2873. doi:10.1093/annonc/mds080
Goldstein LJ, Gray R, Badve S, Childs BH, Yoshizawa C, Rowley S, Shak S, Baehner FL, Ravdin PM, Davidson NE, Sledge GW Jr, Perez EA, Shulman LN, Martino S, Sparano JA (2008) Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol 26(25):4063–4071. doi:10.1200/JCO.2007.14.4501
Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R (2008) Histopathologic variables predict Oncotype DX recurrence score. Mod Pathol 21(10):1255–1261. doi:10.1038/modpathol.2008.54
Geradts J, Bean SM, Bentley RC, Barry WT (2010) The oncotype DX recurrence score is correlated with a composite index including routinely reported pathobiologic features. Cancer Invest 28(9):969–977. doi:10.3109/07357907.2010.512600
Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, Zabaglo L, Mallon E, Green AR, Ellis IO, Howell A, Buzdar AU, Forbes JF (2011) Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol 29(32):4273–4278. doi:10.1200/JCO.2010.31.2835
Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11(10):733–739. doi:10.1038/nrg2825
Lusa L, McShane LM, Reid JF, De Cecco L, Ambrogi F, Biganzoli E, Gariboldi M, Pierotti MA (2007) Challenges in projecting clustering results across gene expression-profiling datasets. J Natl Cancer Inst 99(22):1715–1723. doi:10.1093/jnci/djm216
Baggerly KA, Coombes KR, Neeley ES (2008) Run batch effects potentially compromise the usefulness of genomic signatures for ovarian cancer. J Clin Oncol 26(7):1186–1187; author reply 1187–1188. doi:10.1200/JCO.2007.15.1951
Wang D, Cheng L, Zhang Y, Wu R, Wang M, Gu Y, Zhao W, Li P, Li B, Wang H, Huang Y, Wang C, Guo Z (2012) Extensive up-regulation of gene expression in cancer: the normalised use of microarray data. Mol BioSyst 8(3):818–827. doi:10.1039/c2mb05466c
Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA (2012) Revisiting global gene expression analysis. Cell 151(3):476–482. doi:10.1016/j.cell.2012.10.012
Eddy JA, Sung J, Geman D, Price ND (2010) Relative expression analysis for molecular cancer diagnosis and prognosis. Technol Cancer Res Treat 9(2):149–159
Xu L, Tan AC, Winslow RL, Geman D (2008) Merging microarray data from separate breast cancer studies provides a robust prognostic test. BMC Bioinform 9:125. doi:10.1186/1471-2105-9-125
Russnes HG, Navin N, Hicks J, Borresen-Dale AL (2011) Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest 121(10):3810–3818. doi:10.1172/JCI57088
Bertos NR, Park M (2011) Breast cancer—one term, many entities? J Clin Invest 121(10):3789–3796. doi:10.1172/JCI57100
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. doi:10.1038/nature11412
Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, Gonzalez-Angulo AM, Mills GB, Miller WR, Wu H, Shyr Y, Arteaga CL (2011) A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clin Cancer Res 17(7):2024–2034. doi:10.1158/1078-0432.CCR-10-2567
Louie MC, McClellan A, Siewit C, Kawabata L (2010) Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol Cancer Res 8(3):343–352. doi:10.1158/1541-7786.MCR-09-0395
Fox EM, Miller TW, Balko JM, Kuba MG, Sanchez V, Smith RA, Liu S, Gonzalez-Angulo AM, Mills GB, Ye F, Shyr Y, Manning HC, Buck E, Arteaga CL (2011) A kinome-wide screen identifies the insulin/IGF-I receptor pathway as a mechanism of escape from hormone dependence in breast cancer. Cancer Res 71(21):6773–6784. doi:10.1158/0008-5472.CAN-11-1295
Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A (2010) FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 70(5):2085–2094. doi:10.1158/0008-5472.CAN-09-3746
Musgrove EA, Sutherland RL (2009) Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9(9):631–643. doi:10.1038/nrc2713
Zhou X, Shi T, Li B, Zhang Y, Shen X, Li H, Hong G, Liu C, Guo Z (2013) Genes dysregulated to different extent or oppositely in estrogen receptor-positive and estrogen receptor-negative breast cancers. PLoS One 8(7):e70017
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(Database issue):D991–D995. doi:10.1093/nar/gks1193
Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, Kurbatova N, Malone J, Mani R, Mupo A, Pedro Pereira R, Pilicheva E, Rung J, Sharma A, Tang YA, Ternent T, Tikhonov A, Welter D, Williams E, Brazma A, Parkinson H, Sarkans U (2013) ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res 41(Database issue):D987–D990. doi:10.1093/nar/gks1174
Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacon JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF (2011) A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 305(18):1873–1881. doi:10.1001/jama.2011.593
Garcia-Becerra R, Santos N, Diaz L, Camacho J (2012) Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci 14(1):108–145. doi:10.3390/ijms14010108
Osborne CK, Schiff R (2011) Mechanisms of endocrine resistance in breast cancer. Annu Rev Med 62:233–247. doi:10.1146/annurev-med-070909-182917
Deb K, Pratap A, Agarwal S, Meyarivan T (2002) A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans Evol Comput 6(2):182–197
Kattan MW (2004) Evaluating a new marker’s predictive contribution. Clin Cancer Res 10(3):822–824
Moskowitz CS, Pepe MS (2004) Quantifying and comparing the accuracy of binary biomarkers when predicting a failure time outcome. Stat Med 23(10):1555–1570. doi:10.1002/sim.1747
Therneau TM, Grambsch PM (2000) Modeling survival data: extending the cox model. Springer, New York
Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V, Abraham M, Medeiros Alencar VH, Badran A, Bonfill X, Bradbury J, Clarke M, Collins R, Davis SR, Delmestri A, Forbes JF, Haddad P, Hou MF, Inbar M, Khaled H, Kielanowska J, Kwan WH, Mathew BS, Mittra I, Muller B, Nicolucci A, Peralta O, Pernas F, Petruzelka L, Pienkowski T, Radhika R, Rajan B, Rubach MT, Tort S, Urrutia G, Valentini M, Wang Y, Peto R (2013) Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 381(9869):805–816. doi:10.1016/S0140-6736(12)61963-1
Jin H, Tu D, Zhao N, Shepherd LE, Goss PE (2012) Longer-term outcomes of letrozole versus placebo after 5 years of tamoxifen in the NCIC CTG MA.17 trial: analyses adjusting for treatment crossover. J Clin Oncol 30(7):718–721. doi:10.1200/JCO.2010.34.4010
Colleoni M, Giobbie-Hurder A, Regan MM, Thurlimann B, Mouridsen H, Mauriac L, Forbes JF, Paridaens R, Lang I, Smith I, Chirgwin J, Pienkowski T, Wardley A, Price KN, Gelber RD, Coates AS, Goldhirsch A (2011) Analyses adjusting for selective crossover show improved overall survival with adjuvant letrozole compared with tamoxifen in the BIG 1-98 study. J Clin Oncol 29(9):1117–1124. doi:10.1200/JCO.2010.31.6455
Cuzick J, Sestak I, Baum M, Buzdar A, Howell A, Dowsett M, Forbes JF (2010) Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol 11(12):1135–1141. doi:10.1016/S1470-2045(10)70257-6
Dowsett M, Cuzick J, Ingle J, Coates A, Forbes J, Bliss J, Buyse M, Baum M, Buzdar A, Colleoni M, Coombes C, Snowdon C, Gnant M, Jakesz R, Kaufmann M, Boccardo F, Godwin J, Davies C, Peto R (2010) Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol 28(3):509–518. doi:10.1200/JCO.2009.23.1274
Dubsky PC, Jakesz R, Mlineritsch B, Postlberger S, Samonigg H, Kwasny W, Tausch C, Stoger H, Haider K, Fitzal F, Singer CF, Stierer M, Sevelda P, Luschin-Ebengreuth G, Taucher S, Rudas M, Bartsch R, Steger GG, Greil R, Filipcic L, Gnant M (2012) Tamoxifen and anastrozole as a sequencing strategy: a randomized controlled trial in postmenopausal patients with endocrine-responsive early breast cancer from the Austrian Breast and Colorectal Cancer Study Group. J Clin Oncol 30(7):722–728. doi:10.1200/JCO.2011.36.8993
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant numbers 81071646 and 91029717 to ZG, 81201822 to YG]; and the Research Fund for the Doctoral Program of Higher Education of China [20112307110011 to ZG]. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or decision to publish.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, X., Li, B., Zhang, Y. et al. A relative ordering-based predictor for tamoxifen-treated estrogen receptor-positive breast cancer patients: multi-laboratory cohort validation. Breast Cancer Res Treat 142, 505–514 (2013). https://doi.org/10.1007/s10549-013-2767-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2767-8