Abstract
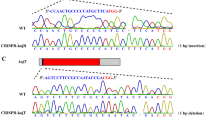
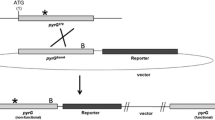
Previously we reported that double disruption of the proteinase genes (tppA and pepE) improved heterologous protein production by Aspergillus oryzae (Jin et al. Appl Microbiol Biotechnol 76:1059–1068, 2007). Since A. oryzae has 134 protease genes, the number of auxotrophy in a single host is limited for multiple disruptions of many protease genes. In order to rapidly perform multiple gene disruptions in A. oryzae, we generated the marker recycling system in highly efficient gene-targeting background. A. oryzae ligD gene homologous to Neurospora crassa mus-53 gene involved in nonhomologous chromosomal integration was disrupted, followed by disruption of the pyrG gene for uridine/uracil auxotroph. We further performed successive rounds of gene disruption (tppA and pepE) by the pyrG marker with high gene-targeting efficiency allowed by the ΔligD background. After each disruption process the pyrG marker was excised by the direct repeats consisting of ~300 bp upstream flanking region of the target gene, resulting in no residual ectopic/foreign DNA fragments in the genome. Consequently, we succeeded to breed the double proteinase gene disruptant (ΔtppA ΔpepE) applicable to further sequential gene disruptions in A. oryzae.




Similar content being viewed by others
References
Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346
d’Enfert C (1996) Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr Genet 30:76–82
Forment JV, Ramón D, MacCabe AP (2006) Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr Genet 50:217–224
Ishibashi K, Suzuki K, Ando Y, Takakura C, Inoue H (2006) Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc Natl Acad Sci USA 103:14871–14876
Ito K, Asakura T, Morita Y, Nakajima K, Koizumi A, Shimizu-Ibuka A, Masuda K, Ishiguro M, Terada T, Maruyama J, Kitamoto K, Misaka T, Abe K (2007) Microbial production of sensory-active miraculin. Biochem Biophys Res Commun 360:407–411
Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K (2004) Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239:79–85
Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K (2007) Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl Microbiol Biotechnol 76:1059–1068
Kimura S, Maruyama J, Takeuchi M, Kitamoto K (2008) Monitoring global gene expression of proteases and improvement of human lysozyme production in nptB gene disruptant of Aspergillus oryzae. Biosci Biotechnol Biochem 72:499–505
Kitamoto K (2002) Molecular biology of the Koji molds. Adv Appl Microbiol 51:129–153
Machida M, Asai K, Sano M et al (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol (in press)
Nakajima K, Asakura T, Maruyama J, Morita Y, Oike H, Shimizu-Ibuka A, Misaka T, Sorimachi H, Arai S, Kitamoto K, Abe K (2006) Extracellular production of neoculin, a sweet-tasting heterodimeric protein with taste-modifying activity, by Aspergillus oryzae. Appl Environ Microbiol 72:3716–3723
Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH (2006) Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol 43:54–64
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101:12248–12253
Takahashi T, Masuda T, Koyama Y (2006) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics 275:460–470
Tsuchiya K, Nagashima T, Yamamoto Y, Gomi K, Kitamoto K, Kumagai C, Tamura G (1994) High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci Biotechnol Biochem 58:895–899
Acknowledgements
We thank to Yukiko Oshima for experimental help. This study was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary Fig. 1
Southern analysis of the ligD disruptants. The genomic DNAs were digested with BamHI and SmaI. The boxed regions are the flanking regions used for disruption of the ligD gene. Four strains were analyzed, confirming that the strains (lanes 1 and 3 in the panels) exhibited the expected band pattern for disruption of the ligD gene. “P” represents the parental strain (NSAR1) (PDF 79 kb)
Supplementary Fig. 2
Southern analysis of the pyrG disruptant. The genomic DNAs were digested with PstI and XhoI. The flanking regions used for disruption of the pyrG gene are boxed. All 4 strains analyzed in the panel (lanes 1 to 4) exhibited the expected band pattern for disruption of the pyrG gene. “P” represents the parental strain (NSR-ΔlD2) (PDF 182 kb)
Rights and permissions
About this article
Cite this article
Maruyama, JI., Kitamoto, K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae . Biotechnol Lett 30, 1811–1817 (2008). https://doi.org/10.1007/s10529-008-9763-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9763-9