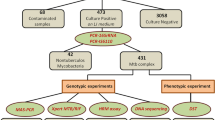
We developed a protocol for detection of mutations in the pncA gene associated with M. tuberculosis resistance to pyrazinamide by analyzing melting curves of 7 overlapping amplicons with artificial heteroduplex formation (H-HRM) formed by co-amplification of wild-type DNA and test DNA and compared its efficiency and robustness with those of classical HRM analysis. Using HRM and H-HRM, we analyzed 35 PZAR DNA isolates carrying mutations in the pncA gene, 3 PZAR isolates without mutations in the pncA gene, and 20 PZAS isolates without mutations in the pncA gene were analyzed. The sensitivity and specificity of HRM for detection of mutations in the pncA gene were moderate: 88.57% (CI 73.26%-96.80%) and 82.61% (CI 61.22%-95.05%), respectively. The sensitivity of the H-HRM test was 97.14% (CI 85.08%-99.93%) and specificity was 95.65% (CI 78.05%-99.89%), with a significant improvement in accuracy — 96.55% vs. 93.85% for HRM. In general, despite addition stage of equalizing the concentrations of the test and control mycobacterial DNA, H-HRM showed greater stability and reproducibility at standard settings of the melting curve analysis software.
Similar content being viewed by others
References
Filipenko ML, Dymova MA, Khrapov EA, Boyarskikh UA, Petrenko TI, Cherednichenko AG, Kozhamkulov UA, Ramankulov EM. Method for detection of rifampicin resistant Mycobacterium tuberculosis isolates. Vestn. Novosibirsk. Gos. Univer. Ser.: Biol. Klin. Med. 2012;10(2):101-106. Russian.
Chang KC, Yew WW, Zhang Y. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis: a systematic review with meta-analyses. Antimicrob. Agents Chemother. 2011;55(10):4499-4505.
Choi GE, Lee SM, Yi J, Hwang SH, Kim HH, Lee EY, Cho EH, Kim JH, Kim HJ, Chang CL. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 2010;48(11):3893-3898.
Hoek KG, Gey van Pittius NC, Moolman-Smook H, Carelse-Tofa K, Jordaan A, van der Spuy GD, Streicher E, Victor TC, van Helden PD, Warren RM. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 2008;46(4):1369-1373.
Morlock GP, Crawford JT, Butler WR, Brim SE, Sikes D, Mazurek GH, Woodley CL, Cooksey RC. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2000;44(9):2291-2295.
Nagai Y, Iwade Y, Hayakawa E, Nakano M, Sakai T, Mitarai S, Katayama M, Nosaka T, Yamaguchi T. High resolution melting curve assay for rapid detection of drug-resistant Mycobacterium tuberculosis. J. Infect. Chemother. 2013;19(6):1116-1125.
Nurwidya F, Handayani D, Burhan E, Yunus F. Molecular diagnosis of tuberculosis. Chonnam. Med. J. 2018;54(1):1-9.
Pholwat S, Liu J, Stroup S, Gratz J, Banu S, Rahman SM, Ferdous SS, Foongladda S, Boonlert D, Ogarkov O, Zhdanova S, Kibiki G, Heysell S, Houpt E. Integrated microfluidic card with TaqMan probes and high-resolution melt analysis to detect tuberculosis drug resistance mutations across 10 genes. MBio. 2015;6(2). ID e02273. doi: 10.1128/mBio.02273-14.
Pholwat S, Stroup S, Gratz J, Trangan V, Foongladda S, Kumburu H, Juma S. P, Kibiki G, Houpt E. Pyrazinamide susceptibility testing of Mycobacterium tuberculosis by high resolution melt analysis. Tuberculosis (Edinb). 2014;94(1):20-25.
Ramirez-Busby SM, Valafar F. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 2015;59(9):5267-5277.
Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245(2):154-160.
Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents. Chemother. 1997;41(3):540-543.
Tong SY, Giffard PM. Microbiological applications of high-resolution melting analysis. J. Clin. Microbiol. 2012;50(11):3418-3421.
Van Soolingen D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 2001;249(1):1-26.
Zhang Y, Shi W, Zhang W, Mitchison D. Mechanisms of pyrazinamide action and resistance. Microbiol. Spectr. 2013;2(4): 1-12.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Byulleten’ Eksperimental’noi Biologii i Meditsiny, Vol. 168, No. 8, pp. 220-226, August, 2019
Rights and permissions
About this article
Cite this article
Filipenko, M.L., Dymova, M.A., Cherednichenko, A.G. et al. Detection of Mutations in Mycobacterium tuberculosis pncA Gene by Modified High-Resolution Melting Curve Analysis of PCR Products. Bull Exp Biol Med 168, 264–269 (2019). https://doi.org/10.1007/s10517-019-04688-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10517-019-04688-6