Abstract
Hog deer were introduced to Australia in the 1860s, where they have spread across the Gippsland region of Victoria. Due to its status as an introduced species and an important game animal within Victoria, management of the species is complex. Given this complexity, genetic studies can provide important information regarding population structure and diversity which can assist in controlling problematic populations of hog deer, while also ensuring viable game stock in sites managed as game reserves. The aim of this study was to investigate the population genetic structure and diversity of the Victorian hog deer 150 years after introduction using short tandem repeats (STRs). Hog deer samples were collected across 15 sites of differing management regimes in the Gippsland region of Victoria and genotyped for 13 polymorphic STR loci. Up to four distinct genetic clusters were identified across the sites sampled, suggesting that despite low observed genetic diversity, population structure is present across their range. It was also possible to detect evidence of recent translocations among populations. This study suggests that the presence of distinct genetic clusters may enable management of separate genetic units, considering invasive species and game management objectives.
Similar content being viewed by others
Introduction
At least 80 species of non-native vertebrates have become established in Australia following European settlement (Bomford and Hart 2002). Many of these species have since become pests to both agriculture and native biodiversity; for example, red foxes (Vulpes vulpes) and wild dogs (Canis familiaris) cause significant damage to livestock and native fauna (Saunders et al. 2010; Davis et al. 2015), while European rabbits (Oryctolagus cuniculus) negatively affect both native and agricultural pastures (Fleming et al. 2002; Mutze et al. 2016). However, while many introduced species cause severe negative impacts, some species can provide limited benefits. For example, many introduced ungulate species, particularly deer, provide a recreational resource for hunters as food and sport and generate economic activity. Management of these taxa can therefore be complex, as considerations of invasive species control and game management may need to be taken into account concurrently.
Six deer species have become established across Australia during the nineteenth century following efforts by Acclimatisation Societies to provide sport for the colonies and boost the aesthetics of the landscape (Moriarty 2004). These species include fallow deer (Dama dama), red deer (Cervus elaphus), sambar deer (Rusa unicolor), Javan rusa deer (Rusa timorensis), chital (Axis axis), and hog deer (Axis porcinus), with at least one of these species present in each State or Territory in Australia. In Victoria, management of deer is complex. All six established wild deer species are considered protected wildlife, requiring permits to control problematic populations of deer, and to hunt the species recreationally. Deer causing damage on public land can be controlled under an Authority to Control Wildlife Permit, and all deer species except hog deer can be destroyed without a permit on private land (Davis et al. 2016). Sambar deer are additionally listed as a ‘potentially threatening process’ under the Flora and Fauna Guarantee Act 1988. All established deer species are also declared game and can be hunted with a game license. Recreational hunting in Victoria provides a significant amount of revenue and economic benefits, with a gross contribution of $356 million AUD and 3138 jobs in 2019 across the State; of this, $201 million AUD is generated by deer hunting alone (Department of Jobs, Precincts and Regions 2020).
Hog deer were introduced to Victoria in the 1860s from an initial founder group of only 15 individuals (Mayze and Moore 1990) and have since grown to encompass a distribution of 2336 km2 across the Gippsland region of Victoria (Fig. 1) (Forsyth et al. 2016). Across this range, management can involve culling to remove the species from Wilsons Promontory National Park to restrictions on hunting to ensure a sustainable population on nearby Sunday Island (Fig. 2). Several game reserves across mainland Victoria also exist to accommodate hog deer hunting, with balloted hunting seasons implemented for Blond Bay State Game Reserve, Boole Poole Peninsula, and Sunday Island to achieve management objectives (Fig. 2) (Salmon 2016). Hog deer are the only deer species in Victoria with a prescribed open season, with hunting restricted to April and only one male and one female harvested per hunter during this open season. These restrictions are in place due to a combination of previous population declines of the species in Victoria when bag limits were not imposed (Taylor 1971; Mayze and Moore 1990), and the possibility that the Victorian population of hog deer may be important for global conservation of the species. In the past, management of hog deer has involved translocations across Victoria in an effort to boost local population numbers. Several deer from Snake Island were translocated to Dutson Downs in the 1970s, while deer from Sunday Island and Serendip Wildlife Reserve near Geelong were released at Blond Bay State Game Reserve in the 1980s (Fig. 1) (Mayze and Moore 1990; Scroggie et al. 2012). While the current population is believed to be derived from the release of 12 deer at Opossum Creek near Port Welshpool in 1865, other releases of hog deer also occurred in the nineteenth century. Eight deer were released near Gembrook in 1871, while a single male and female pair was each released near La Trobe River at Sale, and Homewood (Fig. 1) (Mayze and Moore 1990). Hog deer are no longer present near Gembrook and Homewood, and there is some uncertainty if the pair released near Sale persisted or if the deer from the initial release at Opossum Creek have naturally dispersed to this area. Previous research suggests that the number of individuals released in a single introduction is a strong indicator of establishment success in ungulates, with all releases involving fewer than four individuals deemed to have failed in Victoria (Forsyth et al. 2004). Hog deer were also released at Snake Island in the 1860s (Wednesday, September 4, 1867 1867); however, very little is known of how many deer were released and if the population present on the island today is descended from this introduction. Today, an estimated 3000 hog deer are present throughout Gippsland, however, this is likely to be an underestimation of total abundance as contributions from private land were not considered (Ramsey et al. 2019).
Hog deer release and translocation sites across Victoria, Australia. Green circles indicate sites where hog deer were released by the Acclimatisation Society, with information of the release year and the number of males and females released; orange circles indicate sites involved in translocations with arrows indicating the direction of movement. Blue shaded areas show the overall hog deer distribution across Gippsland, Victoria, as taken from the Atlas of Living Australia (Atlas of Living Australia 2019)
Hog deer sampling sites in Victoria, Australia. Sample sites are (1) Yanakie, (2) Wilsons Promontory National Park, (3) Snake Island, (4) Sunday Island, (5) Gelliondale, (6) Tarraville, (7) Stratford, (8) Perry Bridge, (9) Clydebank, (10) Lake Coleman, (11) Lake Reeve, (12) Loch Sport, (13) Blond Bay, (14) Bengworden, and (15) Boole Poole. Blue shaded regions indicate the overall hog deer distribution across Gippsland, Victoria, as taken from the Atlas of Living Australia (Atlas of Living Australia 2019). Sites 1–6 are considered the western distribution of hog deer, while sites 7–15 are considered the eastern distribution
The historical releases and movement of hog deer across Victoria in the nineteenth and twentieth centuries are well documented; however, little is known about the contemporary population regarding dispersal, and how the introductions, translocations, and hunting of deer have affected the genetics of the species. Although reportedly sedentary in their native range (Dhungel and O’Gara 1991), hog deer have spread across Gippsland following their initial release with a single breeding population assumed (Forsyth et al. 2016); however, it is difficult to ascertain how much of this dispersal was aided by additional releases and translocations. Regardless, some instances of long-distance natural movements of up to 70 km have been observed, with males dispersing further than females (Mayze and Moore 1990). While it is thought that the hog deer range won’t expand further in Gippsland via natural dispersal as the species is limited by biophysical factors such as unsuitable habitats (Forsyth et al 2016), human mediated movement of hog deer may result in range expansions. Illegal translocations of deer by hunters or deliberate releases/escapees from farms may artificially increase the hog deer distribution in Australia; however, only a very small proportion of deer farms contain hog deer (Shapiro 2010), so it is unknown how much of a risk this poses. Over half of all wild deer herds in Australia appear to have arisen from illegal translocations (Moriarty 2004), and genetic studies have been able to successfully detect illegal translocation of deer in Europe (Frantz et al. 2006). It may therefore be possible to verify the extent of illegal translocations of hog deer throughout Gippsland to determine if this is a likely risk for further unwanted expansion of the species.
Assessment of the impacts of sustained harvesting in sites managed as game reserves, and historical hybridisation between hog deer and chital are also of interest, particularly in measurements of genetic diversity. Previous research has shown that hog deer hybridised with chital very early in the initial introduction of the species, with all hog deer tested comprising a chital mitochondrial haplotype, and analysis of mitochondrial DNA revealed local populations of hog deer to be monomorphic at the D-loop region (Hill et al. 2019), suggesting diversity is very low in Victorian hog deer. The low levels of diversity at this mitochondrial region are likely due to founder effects and may be reflective of chital diversity at the time of hybridisation rather than hog deer. Although hybridisation is not an ongoing process in the hog deer population as no chital are present in Victoria, the initial hybridisation event may have increased the genetic diversity at the nuclear genome. Hunting regimes may also impact genetic diversity through sustained removal of individuals, particularly males, over many years. However, the severity of hunting impacts is likely to be dependent on the dispersal capability of hog deer, as high dispersal would suggest that more males can readily move into areas where hunting pressure may be more severe.
Previous research has shown that genetic analysis using short tandem repeats (STRs) is effective at identifying invasion pathways, population connectivity, human mediated dispersal, and genetic variation in introduced populations of invasive species (Walker et al. 2003; Rollins et al. 2009; Yue et al. 2010; LaRue et al. 2011), and these markers are likely to be more informative than the mitochondrial markers that have been used in the past for hog deer (Hill et al. 2019). Knowledge of dispersal and illegal translocation of hog deer can assist in invasive species management and will provide insight into the impact that natural dispersal and illegal translocations may have on the management of this species. Likewise, population connectivity is of importance to areas managed as game reserves; if connectivity is low, excessive removal of individuals associated with hunting within these areas may be unsustainable. Levels of genetic diversity will also be of interest for game managers, as populations with low levels of genetic diversity have been previously shown to be linked with reduced reproductive success (Reed and Frankham 2003), increased vulnerability to disease (Spielman et al. 2004), and decreased adaptability to environmental change (Frankham 2005), all of which may result in local extinctions of hog deer in areas open to hunting. While population genetic studies using STRs have been conducted in the past for deer species in Australia, particularly in rusa deer (Webley et al. 2004) and fallow deer (Webley et al. 2007), these studies have largely focused on a few small populations with limited samples, rather than the entire distributions. This investigation is the first to examine population genetics of the entire wild distribution of a deer species in Australia. The aim of this study is to determine the population structure of hog deer across their Victorian distribution and identify the levels of genetic diversity present in the population.
Methods
Sampling and laboratory procedures
Tissue samples (tongue or liver) were collected from wild, free-ranging hog deer between 2008 and 2017, in Victoria, Australia. A total of 15 sites and 231 samples were selected for study, with site distribution across the hog deer range in Victoria (Fig. 2). These samples were predominantly collected by hunters during the hog deer hunting season held in April, while samples from Wilsons Promontory National Park were collected during culls held in 2015–2016 (Table 1). DNA extractions were carried out using a DNeasy Blood and Tissue Kit (Qiagen), following the manufacturer’s instructions. Negative controls were run throughout the extraction process, and DNA was quantified using a Qubit 2.0 Fluorometer (Invitrogen).
A total of 13 polymorphic STRs were utilised in this study (ApoV103, ApoV104, ApoV19, ApoV135, Apo4, ApoV54, ApoV133, Apo7, ApoV118, ApoV145, ApoV109, ApoV94, and ApoV127), amplified using the multiplexes described in Hill et al. (2021). Details of marker amplification steps, genotyping and filtering of multiplexes, and tests for Hardy–Weinberg equilibrium and linkage disequilibrium can be found in Hill et al. (2021). Samples that failed to amplify two or more loci were removed, leaving 224 samples for further analysis. Markers comprising a null allele frequency above 5% across all samples as reported in Hill et al. (2021) (ApoV19, ApoV104, ApoV94, ApoV145) were further investigated to determine how the presence of null alleles may affect further analyses, by comparing results derived from a null allele corrected dataset and the uncorrected dataset (Supplementary Material 1). From these analyses, it was determined that null alleles at these four markers were not consistently identified across sites, and comparisons of analyses between a null allele corrected dataset and uncorrected dataset did not reveal significant changes to results; therefore, the uncorrected dataset was retained for further analysis.
Data analysis
The program Colony 2.0.6.4 was used to assess kinship between individuals (Jones and Wang 2010), to identify potential dispersal events through kin, and to identify closely related individuals to be removed from the final dataset, as the inclusion of close relatives can affect the accuracy of analyses of diversity and connectivity. Polygamy in both males and females, inbreeding, and no sibship prior or candidate parents were assumed. The pairwise full likelihood combined method for analysis was chosen with high precision and updating allele frequencies, and a 0.005 allele dropout rate and false allele rate per locus were also assumed. Colony 2.0.6.4 was then run five times to ensure convergence of results. A total of 14 sibling pairs with a probability above 0.9 were identified, with some individuals appearing in multiple sibling pairs; a total of 11 individuals were removed from the dataset, leaving 213 individuals for further analysis.
To identify the number of genetically distinct populations within the dataset, several analyses to assess population structure were undertaken. Isolation by distance was investigated via Moran’s eigenvectors, implemented in the R package memgene 1.0.1 (Galpern et al. 2014); this R package is advantageous as it is able to assess fine-scale levels of genetic differentiation. These calculations were implemented for the full dataset, as well as splitting the samples in a western and eastern distribution and running these separately to assess any fine-scale isolation by distance. Samples were split according to a west/east distribution (western sites 1–6, eastern sites 7–15 as shown in Fig. 2) as it was assumed that fine-scale genetic structure would exist between these areas based on the large distance between eastern sites and western sites. Wright’s pairwise FST was calculated between each site using FSTAT 2.9.3.2 (Goudet 1995). The programs Structure 2.3.4 and Tess 2.3 were utilised to compare any population structure detected across the dataset, and spatial coordinate information for each sample was included in Tess to assist in assigning genetic clusters (Pritchard et al. 2003; Chen et al. 2007). An admixture model with correlated allele frequencies was assumed in Structure, with an initial burn-in of 300,000 and 700,000 Monte Carlo Markov Chain (MCMC) repetitions, and 15 iterations per K, ranging from K = 1–15 to reflect the number of sites sampled. Tess was run using the CAR admixture model, with a burn-in of 20,000, and a total of 100,000 sweeps, and 15 iterations per K. Best fits for K were determined using the Evanno method in Structure Harvester 0.6.94 (Evanno et al. 2005; Earl and VonHoldt 2012) for the Structure results and using the average Deviance Information Criterion (DIC) in Tess (Supplementary Material 2). To account for differences between best fit for Structure and Tess, multiple Ks were plotted to compare results between both programs. CLUMPP 1.1.2 was used to summarise the 15 iterations of K for Structure and Tess using the Greedy algorithm and 1000 repeats (Jakobsson and Rosenberg 2007). Plots for both Structure and Tess were created using the R packages ggplot2 3.3.5 and cowplot 1.1.1 (Wickham 2011; Wilke 2019).
Based on population structure results from memgene, FSTAT, Structure, and Tess, some sites were combined for demographic and diversity analyses. Combining sites into one population was restricted to sites that showed genetic similarity and were also adjacent to each other; this therefore excluded sites that were genetically similar but disjointed across the landscape, such as Snake Island and Loch Sport. Additionally, island sites were not combined with genetically similar mainland sites, such as Sunday Island with Gelliondale and Tarraville. Gelliondale and Tarraville were combined to form the ‘GelTar’ population (n = 15), Stratford, Clydebank, and Perry Bridge are incorporated into the ‘StPeCl’ population (n = 27), Lake Coleman and Lake Reeve form the ‘Lakes’ population (n = 15), and Blond Bay and Bengworden are combined into the ‘BloBen’ population (n = 19) for further diversity and demographic analyses.
GenAlEx 6.5.1 was used to calculate number of alleles (Na), observed and expected heterozygosity (HO and HE), and generate a private alleles list (PA) for each site in Victoria (Peakall and Smouse 2012). Allelic richness (AR) was additionally calculated for each site using the PopGenReport 3.0.4 package in R (R Core Team 2021; Adamack and Gruber 2014). The effective population size (Ne) for each sampling site was determined using the Linkage Disequilibrium method in NeEstimator2 (Do et al. 2014), with 95% confidence intervals calculated via jackknifing of samples. Relatedness within sampling sites of hog deer was calculated using the related package in R 3.5.3 (R Core Team 2021; Pew et al. 2015). The ‘compareestimators’ function was used to test the performance of four relatedness models, and similar correlation coefficients between observed and expected values were found (wang = 0.691, lynchli = 0.695, lynchrd = 0.666, quellergt = 0.668), so the lynchli method was selected for further analysis as it comprised the highest correlation coefficient (Li et al. 1993). The ‘grouprel’ function was used to analyse relatedness within sites, using 1000 iterations.
Results
Colony identified 14 full sibling pairs with probabilities greater than 0.9, with no half sibling pairs identified above this threshold. Seven full sibling pairs were within the same sites, with four of these pairs from Snake Island (Supplementary Material 3). Seven pairs were between sites, with three of these pairs between Snake Island and Loch Sport, two pairs between Lake Coleman and Boole Poole, one pair between Tarraville and Boole Poole, and one pair between Tarraville and Lake Coleman. All Loch Sport and Snake Island sibling pairs were born within a 2-year period (2013–2014), and all were collected in 2017.
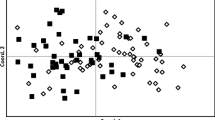
Memgene results showed that the amount of genetic variation explained by spatial patterns was low for the entire dataset (R2Adj = 0.09) as well as the western and eastern sites only (R2Adj = 0.077 and R2Adj = 0.075, respectively) (Fig. 3). The first Moran’s eigenvector maps (MEM) score for the full dataset showed a clear distinction between the western and eastern sites; however, Tarraville, a western site, was shown to be more similar to the eastern sites rather than other western sites. This first variable accounted for 62% of the variation observed (Fig. 3a). Within the western sites only, the first MEM score showed a separation of Snake Island and Wilsons Promontory to Yanakie, Sunday Island, Gelliondale, and Tarraville (Fig. 3b), with this MEM variable explaining 80% of the spatial variation. Tarraville was not identified as being different to the other sites when considering western sites only. The first MEM score for the eastern sites showed that Loch Sport and Boole Poole were separate from all other eastern sites, with this variable explaining 82% of the variation (Fig. 3c).
Visualisations of the first MEM variables for a the entire hog deer dataset, b the western Victorian sites, and c the eastern Victorian sites. Black and white circles indicate positive and negative axis score values, respectively (i.e., different genetic groups), and circle size indicates genetic differences within the axis scores (i.e., circles of similar sizes are more genetically similar than circles of different sizes)
Pairwise FST results show that a majority of pairwise comparisons are significantly different (Table 2). In particular, Wilsons Promontory, Snake Island, and Boole Poole appeared to be highly significantly different from almost all other sites sampled. All comparisons to Snake Island showed a high degree of significance with an adjusted p-value of 0.000476, with the exception of Loch Sport which was only significant below 0.05. Within the western sites, only pairwise values between Yanakie and Gelliondale, and Gelliondale and Tarraville were non-significant; in contrast, many of the pairwise comparisons from the eastern sites were non-significant. When comparing pairwise FSTs between the western and eastern sites, a majority of these were again significantly different; however, comparisons to Blond Bay, Lake Reeve, Gelliondale, and Tarraville led a majority of the non-significant values observed between western and eastern sites.
The best fit for K was determined to be K = 2 for Structure and K = 4 for Tess, with outputs of K from 2 to 4 plotted from each program. The genetic structure detected in Structure and Tess appears to be relatively similar in terms of the relationships between clusters; however, the addition of geographic coordinates in Tess has provided a greater clarity at assigning sites to clusters (Fig. 4). Snake Island appears to be highly genetically distinct from all other sites; however, some closely related individuals were also detected in the Stratford and Loch Sport sites. This pattern is observed through K 2–4 in both the Structure and Tess plots. Boole Poole also appears to be genetically distinct, with some individuals from Tarraville, Lake Coleman, and Lake Reeve also assigned to this cluster; however, this pattern is more apparent in Tess than Structure. At K = 4, results differ between the Structure and Tess outputs; a Wilsons Promontory cluster is apparent in the Tess plot, with individuals from Yanakie, Sunday Island, Gelliondale, and Tarraville also assigned to this group. The fourth cluster is predominant across eastern sites Stratford, Clydebank, Perry Bridge, Lake Coleman, Lake Reeve, Blond Bay, and Bengworden; however, it is also present in western sites Yanakie, Sunday Island, Gelliondale, and Tarraville. In the Structure K = 4 plot, these two clusters are equally spread across the western and eastern sites.
Structure and Tess plots for all samples of hog deer, showing Ks 2–4. Best fit for K was K = 2 for Structure, and K = 4 for Tess. Samples are ordered from west to east across the range of sites sampled across Victoria, Australia (1) Yanakie, (2) Wilsons Promontory National Park, (3) Snake Island, (4) Sunday Island, (5) Gelliondale, (6) Tarraville, (7) Stratford, (8) Perry Bridge, (9) Clydebank, (10) Lake Coleman, (11) Lake Reeve, (12) Loch Sport, (13) Blond Bay, (14) Bengworden, and (15) Boole Poole
Following analyses of population structure, genetically similar sites (1) Gelliondale and Tarraville, (2) Stratford, Clydebank, and Perry Bridge, (3) Lake Coleman and Lake Reeve, and (4) Blond Bay and Bengworden were combined for analyses of genetic diversity and relatedness within sample sites.
The overall observed heterozygosity (HO) recorded in Victoria was 0.412; however, minimal variation was shown between sites, with values between 0.368 and 0.449 (Table 3). Only three sites recorded a HO below 0.4, Wilsons Promontory (0.368), Snake Island (0.382), and Loch Sport (0.385). Allelic richness (AR) was also similar across sites, with values between 1.983 and 2.3, with Wilsons Promontory comprising the lowest allelic richness. A single private allele was recorded each at Sunday Island, Boole Poole, and the combined StPeCl population, with the individual comprising a private allele from the StPeCl population being collected from Clydebank. Relatedness within each site sampled ranged from − 0.203 to 0.267, indicating a wide variation of relatedness across Victoria (Table 3). Boole Poole and Wilsons Promontory comprised the two highest relatedness levels of 0.267 and 0.213, respectively. Estimates of effective population size (Ne) were low across the entire distribution sampled, with a total Ne value of 39 individuals (95% C.I. = 26–59).
Discussion
This study represents the first population genetic analysis of the entire wild distribution of an introduced deer species in Australia, which has successfully detected population structure in what was previously thought to be a single breeding population. Genetic diversity was relatively similar across sites and consistent with other measures of STR diversity in introduced deer species in Australia, albeit with different marker sets (Webley et al. 2004, 2007). Diversity may have been expected to be higher in the population due to historic hybridisation with chital, with hybridisation typically leading to increased heterozygosity; however, hybridisation is not an ongoing process within this population, with the initial hybridisation event occurring very early in the introduction of the species. Isolation by distance was evident between the western and eastern sites, with the exception of one western site being more closely related to eastern sites, and pairwise FST showed weaker genetic structure present in eastern sites than in western sites. Evidence of translocations were also observed between several sites. The detection of population structure across the hog deer population in Victoria indicates that management of separate units can be conducted, considering invasive species and game management imperatives.
Genetic structure
Significant genetic structure was detected across the Victorian distribution of hog deer, demonstrating that multiple distinct breeding populations exist. Up to four distinct genetic clusters were identified using Structure and Tess, and pairwise FST revealed genetic structure between multiple sites. The addition of geographic data integrated in Tess appears to have assisted in elucidating additional fine-scale structure present across the hog deer range. The ‘Snake Island’ cluster, comprising all samples from Snake Island as well as individuals from Stratford and Loch Sport, showed very strong genetic differentiation to all other sampling sites in Victoria. No private alleles were observed in the Snake Island population, and previous research did not reveal any differences in mitochondrial or nuclear sequences when compared to other populations (Hill et al. 2019). This observed genetic difference may be due to Snake Island being one of the initial hog deer release sites, undergoing a separate founder event and consequent genetic drift due to the long-lasting isolation from the mainland populations.
Significant genetic structure was also detected at Wilsons Promontory and Boole Poole, suggesting that these sites may be genetically isolated from other areas. These two sites represent the most western and eastern sites sampled in this study and represent the range edges of the species in Victoria. Populations at range edges of a species are expected to comprise lower genetic diversity and higher levels of genetic structure; however, these patterns are not always consistent (Wagner et al. 2012; Assis et al. 2013; Zigouris et al. 2012). In the present study, genetic diversity was relatively uniform across all sites; however, within-group relatedness was highest at Wilsons Promontory and Boole Poole, which may further support possible range edge effects. Alternatively, these results may also be reflective of hunting pressure, particularly at Boole Poole which is managed as a game reserve.
In many studies investigating genetic structure across introduced populations, the structure detected is typically attributed to multiple founding events (Lecis et al. 2008; Brown and Stepien 2009; Zalewski et al. 2010; Bai et al. 2012). Given that the Victorian population of hog deer has likely arisen from a single founding event, genetic drift may alternatively be responsible for much of the structure observed, which is commonly exacerbated by low effective population sizes (Ne) (Harris et al. 2002). Low Ne causes allele frequencies to change at a much faster rate due to genetic drift, which can cause populations to lose genetic diversity and become genetically distinct over a relatively short period of time (Masel 2011). An example of this has been observed in an introduced population of the common wall lizard (Podarcis muralis) in Germany, where strong genetic differentiation was observed at a small spatial scale and attributed to genetic drift, particularly at sites close to the range margin (Schulte et al. 2013). Similar genetic processes may explain the genetic structure observed in hog deer.
Gene flow via translocations and dispersal
Although genetic structure was detected across the hog deer distribution, evidence of gene flow between sites was also observed. Of interest was gene flow detected between Snake Island, Loch Sport, and Stratford, and sites Boole Poole, Lake Coleman, and Tarraville, where full sibling pairs were also observed within these groups. This suggests that gene flow between these groups is relatively recent (within a single generation), and given that these sites are not adjacent, likely suggests recent translocations from Snake Island to Loch Sport, and from Boole Poole to Lake Coleman and Tarraville. Although genetic similarity was also observed between Snake Island and Stratford, no full sibling pairs were detected between these sites; this may represent an older translocation event where it is no longer possible to detect full sibling pairs, or insufficient sampling where siblings may have been missed. While legal translocations of hog deer have been carried out in the past, with a number of deer translocated from Snake Island to Dutson Downs in the 1970s, and deer from Sunday Island to Blond Bay in the 1980s (Bentley 1978; Slee and Young 1986; Mayze and Moore 1990), the inclusion of kinship data allows the distinction between historic and recent gene flow events. Illegal translocations often occur to supplement pre-existing populations or to create new populations (Seddon et al. 2012), and are commonly observed in ungulate species, including red deer (Frantz et al. 2006) and feral pigs (Sus scrofa) (Spencer and Hampton 2005; Frantz et al. 2009). Many of the deer populations present in Australia today are believed to be the result of translocations (Moriarty 2004).
Although hog deer are considered a sedentary species (Dhungel and O’Gara 1991; Taylor 1971), genetic similarity between multiple adjacent sites was observed, particularly across eastern Gippsland. Introduced species have been shown to increase their dispersal capabilities over time in non-native ranges (Phillips et al. 2006; Alford et al. 2009; Lindström et al. 2013). Alternatively, the patterns observed here may represent continued dispersal over time, whereby the gene flow detected has occurred over many sustained dispersal events of hog deer and not a single dispersal event. Sampling design in the present study may also contribute to some of the patterns observed; genetic samples provided in this study were supplied predominantly by recreational hunters who preferentially harvest males (Scroggie et al. 2012), and while efforts were made to include as many samples from females as possible, the sex ratios skew towards more males in the final dataset. In many mammalian species, males are the dispersing sex (Dobson 1982), and anecdotal evidence suggests this may be the case in hog deer (Mayze and Moore 1990). The moderate levels of dispersal observed in this study may therefore reflect male biased sex dispersal, and additional samples from female hog deer are necessary to understand how sex may play a role in the dispersal of this species.
Management implications
The presence of genetically distinct clusters across the hog deer sites sampled in this study suggests that it will be possible to effectively control problematic populations of hog deer. Culling of hog deer has been occurring at Wilsons Promontory National Park since 2015 which has resulted in a decrease in abundance (Game Management Authority 2017), and while some analyses suggest that this site is isolated, there is also some evidence of gene flow from surrounding sites. It was not possible to elucidate the direction of any potential migrations or translocations and given that gene flow to Wilsons Promontory was not conclusive, additional analyses in western Gippsland are necessary to understand fine-scale dispersal across this region and identify potential reinvasion pathways that may affect culling success. The discovery of translocations of hog deer across the landscape suggests that this may be an issue for ongoing invasive species management, either by supplementing pre-existing populations or by extending the hog deer range to new areas. While human mediated movement into Wilsons Promontory is unlikely to be a concern as recreational hunters are not able to hunt in National Parks, the extension of the current hog deer range in Victoria is worrying. However, the data presented here provides a good baseline to identify the origin of any new populations of hog deer that may arise in the future. To further understand fine-scale movements of hog deer across their range, particularly at sites where population control is routinely carried out, analyses of kinship may be a viable option and has been used in the past to assess fine-scale dispersal (Vangestel et al. 2011; Escoda et al. 2017). While kin were not identified at Wilson Promontory National Park in the present study, an increase in the number of samples taken at this site and surrounding areas and the inclusion of additional STR or single nucleotide polymorphism (SNP) markers may elucidate additional familial relationships.
Genetic diversity did not appear to be different in sites managed as game reserves, and although high within-group relatedness observed at Boole Poole, this may be attributed to either range edge effects or hunting pressure and needs to be investigated further. Gene flow was detected between most game reserves and nearby sites, suggesting that deer are able to disperse from these hunting areas. This was observed at Sunday Island, where gene flow appears to be occurring from the island to nearby mainland sites, and Blond Bay, which showed genetic similarities to Bengworden. Intense harvesting has been shown to increase the rate of gene flow in some species as they disperse further distances to find less disturbed habitats (Allendorf et al. 2008). Recent admixture of two genetically distinct groups of Guinea baboon (Papio papio) in Guinea-Bissau has been recognised due to hunting pressures (Da Silva et al. 2014), and an increase of unrelated males in packs of grey wolves (Canis lupus) was attributed to the ease of recruitment of migrant wolves to new areas due to hunting pressure (Jędrzejewska et al. 2005). While this study was able to measure gene flow from sites managed as game reserves, it was not possible to distinguish between dispersal due to increased hunting pressure or natural dispersal rates of the species. Further fine-scale analyses of dispersal from these sites and comparisons to areas with lower hunting rates may be able to elucidate how hunting pressure affects dispersal of the species.
Conclusion
This study represents the first population genetic analysis across the entire distribution of an introduced deer species in Australia. The detection of population structure across the hog deer range in Victoria indicates that management of separate units is possible, and considerations of different management strategies can be applied to the findings of this study. While the data presented here provides a good baseline for inferring the dispersal ability in hog deer, additional studies of fine-scale migration between sites that appear to be genetically similar would be beneficial to determine how regularly movement between sites is occurring. Further attention should also be given to the other deer species present in Australia, particularly for species such as sambar deer, fallow deer, chital, and red deer as their distributions still appear to be expanding.
Availability of data
Raw data has been uploaded to CloudStor and is available here: https://cloudstor.aarnet.edu.au/plus/s/YjgRWf2MHVvixh1.
Code availability
Not applicable.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5(4):384–387
Alford RA, Brown GP, Schwarzkopf L, Phillips BL, Shine R (2009) Comparisons through time and space suggest rapid evolution of dispersal behaviour in an invasive species. Wildlife Res 36(1):23–28
Allendorf FW, England PR, Luikart G, Ryman RPA, N, (2008) Genetic effects of harvest on wild animal populations. Trends Ecol Evol 23(6):327–337
Assis J, Castilho Coelho N, Alberto F, Valero M, Raimondi P, Reed D, Alvares Serrão E (2013) High and distinct range-edge genetic diversity despite local bottlenecks. PLoS ONE 8(7):e68646
Atlas of Living Australia occurrence download at https://biocache.ala.org.au/occurrences/search?q=lsid%3Aurn%3Alsid%3Abiodiversity.org.au%3Aafd.taxon%3A751e6627-f63b-4c1b-911e-f4ad688be569. Accessed 4 Sept 2019
Bai C, Ke Z, Consuegra S, Liu X, Li Y (2012) The role of founder effects on the genetic structure of the invasive bullfrog (Lithobates catesbeianaus) in China. Biol Invasions 14(9):1785–1796
Bentley A (1978) An introduction to the deer of Australia: with special reference to Victoria. The Koetong Trust Service Fund. Victoria, Australia
Bomford M, Hart Q (2002) Non-indigenous vertebrates in Australia. In: Pimental D (ed) Biological invasions: Economic and environmental costs of alien plant, animal, and microbe Species’, pp 25–45
Brown JE, Stepien CA (2009) Invasion genetics of the Eurasian round goby in North America: tracing sources and spread patterns. Mol Ecol 18(1):64–79
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7(5):747–756
Da Silva MF, Godinho R, Casanova C, Minhós T, Sá R, Bruford MW (2014) Assessing the impact of hunting pressure on population structure of Guinea baboons (Papio papio) in Guinea-Bissau. Conserv Genet 15(6):1339–1355
Davis NE, Bennett A, Forsyth DM, Bowman DM, Lefroy EC, Wood SW, Johnson CN (2016) A systematic review of the impacts and management of introduced deer (family Cervidae) in Australia. Wildl Res 43(6):515–532
Davis NE, Forsyth DM, Triggs B, Pascoe C, Benshemesh J, Robley A, Lumsden LF (2015) Interspecific and geographic variation in the diets of sympatric carnivores: dingoes/wild dogs and red foxes in south-eastern Australia. PLoS ONE 10(3):e0120975
Department of Jobs, Precincts and Regions (2020) Economic contribution of recreational hunting in Victoria, final report. RM Consulting Group. Bendigo, Victoria, Australia
Dhungel SK, O’Gara BW (1991) Ecology of the hog deer in Royal Chitwan National Park, Nepal. Wildlife Monographs, pp 3–40
Do C, Waples RS, Peel D, Macbeth G, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14(1):209–214
Dobson FS (1982) Competition for mates and predominant juvenile male dispersal in mammals. Anim Behav 30(4):1183–1192
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Escoda L, González-Esteban J, Gómez A, Castresana J (2017) Using relatedness networks to infer contemporary dispersal: application to the endangered mammal Galemys pyrenaicus. Mol Ecol 26(13):3343–3357
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Fleming P, Croft J, Nicol H (2002) The impact of rabbits on a grazing system in eastern New South Wales. 2. Sheep production. Australian J Experimental Agric 42(7):917–923
Forsyth DM, Duncan RP, Bomford M, Moore G (2004) Climatic suitability, life-history traits, introduction effort, and the establishment and spread of introduced mammals in Australia. Conserv Biol 18(2):557–569
Forsyth DM, Stamation K, Woodford L (2016) Distributions of fallow deer, red deer, hog deer and chital deer in Victoria. Arthur Rylah Institute for Environmental Research Unpublished Client Report for the Biosecurity Branch. Heidelberg, Victoria
Frankham R (2005) Genetics and extinction. Biol Cons 126(2):131–140
Frantz A, Cellina S, Krier A, Schley L, Burke T (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol 46(2):493–505
Frantz AC, Pourtois JT, Heuertz M, Schley L, Flamand MC, Krier A, Burke T (2006) Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol 15(11):3191–3203
Galpern P, Peres-Neto PR, Polfus J, Manseau M (2014) MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol Evol 5(10):1116–1120
Game Management Authority (2017) Wilsons Promontory National Park hog deer control program. Game Management Authority. Victoria, Australia
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Harris RB, Wall WA, Allendorf FW (2002) Genetic consequences of hunting: what do we know and what should we do? Wildlife Soc Bullet 634–643
Hill E, Linacre A, Toop S, Murphy N, Strugnell J (2019) Widespread hybridization in the introduced hog deer population of Victoria, Australia, and its implications for conservation. Ecol Evol 9(18):10828–10842
Hill E, Linacre A, Toop S, Murphy N, Strugnell JM (2021) Development of an STR panel for a non-native population of an endangered species. Mol Biol Rep. https://doi.org/10.1007/s11033-021-06905-w
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806
Jędrzejewski W, Branicki W, Veit C, MeĐugorac I, Pilot M, Bunevich AN, Okarma H (2005) Genetic diversity and relatedness within packs in an intensely hunted population of wolves Canis lupus. Acta Theriol 50(1):3–22
Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10(3):551–555
LaRue EA, Ruetz CR, Stacey MB, Thum RA (2011) Population genetic structure of the round goby in Lake Michigan: implications for dispersal of invasive species. Hydrobiologia 663(1):71–82
Lecis R, Ferrando A, Ruiz-Olmo J, Mañas S, Domingo-Roura X (2008) Population genetic structure and distribution of introduced American mink (Mustela vison) in Spain, based on microsatellite variation. Conserv Genet 9(5):1149–1161
Li CC, Weeks DE, Chakravarti A (1993) Similarity of DNA fingerprints due to chance and relatedness. Hum Hered 43(1):45–52
Lindström T, Brown GP, Sisson SA, Phillips BL, Shine R (2013) Rapid shifts in dispersal behavior on an expanding range edge. Proc Natl Acad Sci 110(33):13452–13456
Masel J (2011) Genetic drift. Curr Biol 21(20):R837–R838
Mayze RJ, Moore G (1990) The hog deer. Australian Deer Research Foundation, Croydon, Victoria
Moriarty A (2004) The liberation, distribution, abundance and management of wild deer in Australia. Wildl Res 31(3):291–299
Mutze G, Cooke B, Jennings S (2016) Density-dependent grazing impacts of introduced European rabbits and sympatric kangaroos on Australian native pastures. Biol Invasions 18(8):2365–2376
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539
Pew J, Muir PH, Wang J, Frasier TR (2015) related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol Ecol Resour 15(3):557–561
Phillips BL, Brown GP, Webb JK, Shine R (2006) Invasion and the evolution of speed in toads. Nature 439(7078):803–803
Pritchard JK, Wen W, Falush D (2003) Documentation for STRUCTURE software: version 2
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Ramsey DSL, Pacioni C, Hill E (2019) Abundance and population genetics of hog deer (Axis porcinus) in Victoria. Arthur Rylah Institute for Environmental Research Technical Report Series No. 303. Department of Environment, Land, Water and Planning: Heidelberg, Victoria
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17(1):230–237
Rollins LA, Woolnough AP, Wilton AN, Sinclair R, Sherwin WB (2009) Invasive species can’t cover their tracks: using microsatellites to assist management of starling (Sturnus vulgaris) populations in Western Australia. Mol Ecol 18(8):1560–1573
Salmon M (2016) Results of 2016 balloted hunting at Blond Bay State Game Reserve and Boole Poole Peninsula harvest summary report. Game Management Authority. Victoria, Australia
Saunders GR, Gentle MN, Dickman CR (2010) The impacts and management of foxes Vulpes vulpes in Australia. Mammal Rev 40(3):181–211
Schulte U, Veith M, Mingo V, Modica C, Hochkirch A (2013) Strong genetic differentiation due to multiple founder events during a recent range expansion of an introduced wall lizard population. Biol Invasions 15(12):2639–2649
Scroggie MP, Forsyth DM, Brumley AR (2012) Analyses of Victorian hog deer (Axis porcinus) checking station data: demographics, body condition and time of harvest. Arthur Rylah Institute of Environmental Research Technical Report Series, No. 230
Seddon PJ, Strauss WM, Innes J (2012) Animal translocations: what are they and why do we do them. Reintroduct Biol Integr Sci Manage 12(1)
Shapiro S (2010) Deer industry database. Rural Industries Research and Development Corporation, ACT, Australia
Slee K, Young D (1986) Management plan—Blond Bay hog deer project. Melbourne, Australia
Spencer PB, Hampton JO (2005) Illegal translocation and genetic structure of feral pigs in Western Australia. J Wildl Manag 69(1):377–384
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5(4):439–448
Taylor PG (1971) Aspects of the biology of the hog deer (Axis porcinus Zimmerman 1780). (PhD thesis), Monash University, Melbourne
Vangestel C, Mergeay J, Dawson DA, Vandomme V, Lens L (2011) Spatial heterogeneity in genetic relatedness among house sparrows along an urban–rural gradient as revealed by individual-based analysis. Mol Ecol 20(22):4643–4653
Wagner V, Treiber J, Danihelka J, Ruprecht E, Wesche K, Hensen I (2012) Declining genetic diversity and increasing genetic isolation toward the range periphery of Stipa pennata, a Eurasian feather grass. Int J Plant Sci 173(7):802–811
Walker N, Hulme P, Hoelzel A (2003) Population genetics of an invasive species, Heracleum mantegazzianum: implications for the role of life history, demographics and independent introductions. Mol Ecol 12(7):1747–1756
Webley L, Zenger K, English A, Cooper D (2004) Low levels of genetic variation within introduced Javan rusa deer (Cervus timorensis russa) in Australia. Eur J Wildl Res 50(3):137–140
Webley LS, Zenger KR, Hall GP, Cooper DW (2007) Genetic structure of introduced European fallow deer (Dama dama dama) in Tasmania. Australia Eur J Wildlife Res 53(1):40–46
Wednesday September 4, 1867 (4 September 1867) The Argus (Melbourne, Vic. : 1848 - 1957), p. 5. Retrieved from http://trove.nla.gov.au/newspaper/article/5776983/514432
Wickham H (2011) ggplot2. Wiley Interdisciplinary Reviews: Comput Stat 3(2):180–185
Wilke CO (2019) Cowplot: streamlined plot theme and plot annotations for “ggplot2”. R package version 1.0.0
Yue GH, Li J, Bai Z, Wang CM, Feng F (2010) Genetic diversity and population structure of the invasive alien red swamp crayfish. Biol Invasions 12(8):2697–2706
Zalewski A, Michalska-Parda A, Bartoszewicz M, Kozakiewicz M, Brzeziński M (2010) Multiple introductions determine the genetic structure of an invasive species population: American mink Neovison vison in Poland. Biol Cons 143(6):1355–1363
Zigouris J, Dawson FN, Bowman J, Gillett RM, Schaefer JA, Kyle CJ (2012) Genetic isolation of wolverine (Gulo gulo) populations at the eastern periphery of their North American distribution. Conserv Genet 13(6):1543–1559
Acknowledgements
This project was jointly funded by the RFA grant ‘Securing Food, Water and the Environment’ (La Trobe University), and the Victorian Game Management Authority. We would like to thank hunters who provided access to samples, and to Victorian Hog Deer Checking Station operators, Parks Victoria, and Museums Victoria for assisting with the collection of hog deer samples. We would also like to thank Tim Thomas for providing Sunday Island hog deer samples from 2008, and James O’Dwyer for his assistance with related analyses.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This project was jointly funded by the RFA grant ‘Securing Food, Water and the Environment’ (La Trobe University), and the Victorian Game Management Authority.
Author information
Authors and Affiliations
Contributions
EH, AL, ST, NM, and JS contributed to the design of the study; ST and EH arranged collection of Australian samples; EH performed the laboratory procedures; EH, NM, and JS conducted the analyses; EH, NM, and JS led the writing of the manuscript; AL, ST, NM, and JS provided feedback and revisions of the manuscript. All authors contributed equally to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hill, E., Murphy, N., Toop, S. et al. Genetic analysis of hog deer (Axis porcinus) in Victoria, Australia, and its applications to invasive species and game management. Eur J Wildl Res 68, 45 (2022). https://doi.org/10.1007/s10344-022-01592-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-022-01592-9