Abstract
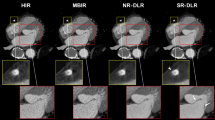
Whole-heart coronary magnetic resonance angiography (WHCMRA) permits the noninvasive assessment of coronary artery disease without radiation exposure. However, the image resolution of WHCMRA is limited. Recently, convolutional neural networks (CNNs) have obtained increased interest as a method for improving the resolution of medical images. The purpose of this study is to improve the resolution of WHCMRA images using a CNN. Free-breathing WHCMRA images with 512 × 512 pixels (pixel size = 0.65 mm) were acquired in 80 patients with known or suspected coronary artery disease using a 1.5 T magnetic resonance (MR) system with 32 channel coils. A CNN model was optimized by evaluating CNNs with different structures. The proposed CNN model was trained based on the relationship of signal patterns between low-resolution patches (small regions) and the corresponding high-resolution patches using a training dataset collected from 40 patients. Images with 512 × 512 pixels were restored from 256 × 256 down-sampled WHCMRA images (pixel size = 1.3 mm) with three different approaches: the proposed CNN, bicubic interpolation (BCI), and the previously reported super-resolution CNN (SRCNN). High-resolution WHCMRA images obtained using the proposed CNN model were significantly better than those of BCI and SRCNN in terms of root mean squared error, peak signal to noise ratio, and structure similarity index measure with respect to the original WHCMRA images. The proposed CNN approach can provide high-resolution WHCMRA images with better accuracy than BCI and SRCNN. The high-resolution WHCMRA obtained using the proposed CNN model will be useful for identifying coronary artery disease.



Similar content being viewed by others
References
World Health Organization. Available at http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death. Accessed 2 September 2018.
Wu W, Liu Z, He X: Learning-based super resolution using kernel partial least squares. Image and Vision Computing 29(6):394–406, 2011
Xian Y, Yang X, Tian Y: Hybrid example-based single image super-resolution. In: International Symposium on Visual Computing, Vol. 9475, 2015, pp. 3–15
Yang CY, Yang MH: Fast direct super-resolution by simple functions. In Proceedings of the IEEE International Conference on Computer Vision:561–568, 2013
Hizukuri A, Nakayama R: Computer-aided diagnosis scheme for determining histological classification of breast lesions on ultrasonographic images using convolutional neural network. Diagnostics 8(3):1–9, 2018
Kumar K, Rao ACS: Breast cancer classification of image using convolutional neural network. In 2018 4th International Conference on Recent Advances in Information Technology (RAIT):1–6, 2018
Dmitriev K, Kaufman AE, Javed AA, Hruban RH, Fishman EK, Lennon AM, Saltz JH: Classification of pancreatic cysts in computed tomography images using a random forest and convolutional neural network ensemble. In: International Conference on Medical mage Computing and Computer-Assisted Intervention 10435, 2017, pp. 150–158
Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, El-Deiry MW, Chen AY, Fei B: Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. Journal of Biomedical Optics 22(6):060503–0601–4, 2017
Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok VCT, Shi L, Heng PA: Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Transactions on Medical Imaging 35(5):1182–1195, 2016
Pereira S, Pinto A, Alves V, Silva CA: Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Transactions on Medical Imaging 35(5):1240–1251, 2016
Park J, Hwang D, Kim KY, Kang SK, Kim YK, Lee JS: Computed tomography super-resolution using deep convolutional neural network. Physics in Medicine and Biology 63(14):145011, 2018
Dong C, Loy CC, He K, Tang X: Image super-resolution using deep convolutional networks. IEEE Transactions on Pattern Analysis and Machine Intelligence 38(2):295–307, 2016
Dong C, Loy CC, He K, Tang X: Learning a deep convolutional network for image super-resolution. In: European Conference on Computer Vision, 2014, pp. 184–199
Umehara K, Ota J, Ishimaru N, Ohno S, Okamoto K, Suzuki T, Shirai N, Ishida T: Super-resolution convolutional neural network for the improvement of the image quality of magnified images in chest radiographs. In Medical Imaging 2017: Image Processing. International Society for Optics and Photonics 10133:101331P1–101331P7, 2017
Magnotta VA, Friedman L: BIRN F: Measurement of signal-to-noise and contrast-to-noise in the fBIRN multicenter imaging study. Journal of Digital Imaging 19(2):140–147, 2006
Dincer HA, Gokcay D: Evaluation of MRIS of the brain with respect to SNR, CNR and GWR in young versus old subjects. In Signal Processing and Communications Applications Conference (SIU):1079–1082, 2014
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP: Image quality assessment: From error visibility to structural similarity. IEEE Transactions on Image Processing 13(4):600–612, 2004
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kobayashi, H., Nakayama, R., Hizukuri, A. et al. Improving Image Resolution of Whole-Heart Coronary MRA Using Convolutional Neural Network. J Digit Imaging 33, 497–503 (2020). https://doi.org/10.1007/s10278-019-00264-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00264-6