Abstract
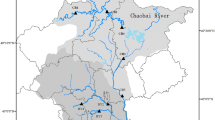
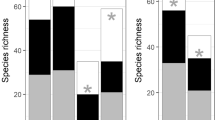
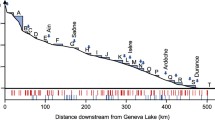
In recent years, biodiversity loss has become one of the most serious environmental issues worldwide, especially in aquatic ecosystems. To avoid diversity loss, it is necessary to monitor biological communities, and environmental DNA (eDNA) metabarcoding has been developed as a rapid, noninvasive, and cost-effective method for aquatic biodiversity monitoring. Although this method has been applied to various environments and taxa, a detailed assessment of the efficient sampling methods for monitoring is still required. In this study, we explored eDNA metabarcoding sampling methods for fish at a single site to maximize the number of detected species using realistic effort in a natural, small river. We considered the following three parameters: sample type (water or sediment), sample position at a site (right and left shore and center of the river), and water volume (10–4000 mL). The results suggested that the number of detected species from sedimentary eDNA was equivalent to that from aqueous eDNA, although the species composition was different. The number of detected species could be saturated by collecting a 1000 mL water sample, regardless of sampling position within a survey site. However, sedimentary eDNA showed a spatially heterogeneous species composition between sampling positions within a survey site despite the short distance (5 m) between positions, without apparent differences in physical properties such as velocity and sediment particle distribution. By completing eDNA biodiversity monitoring of fish with 1000 mL water samples across the whole river, we detected more fish species than in previous traditional surveys conducted at the same sites. Thus, the aqueous eDNA metabarcoding method is as efficient as traditional surveys, while sedimentary eDNA metabarcoding could complement the results of aqueous eDNA metabarcoding.







Similar content being viewed by others
References
Andruszkiewicz EA, Sassoubre LM, Boehm AB (2017) Persistence of marine fish environmental DNA and the influence of sunlight. PLoS ONE 12(9):e0185043. https://doi.org/10.1371/journal.pone.0185043
Barnes MA, Turner CR, Jerde CL et al (2014) Environmental conditions influence eDNA persistence in aquatic systems. Environ Sci Technol 48:1819–1827. https://doi.org/10.1021/es404734p
Bessey C, Jarman SN, Berry O et al (2020) Maximizing fish detection with eDNA metabarcoding. Environ DNA. https://doi.org/10.1002/edn3.74
Bista I, Carvalho GR, Walsh K et al (2017) Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat Commun 8:14087. https://doi.org/10.1038/ncomms14087
Butchart SHM, Walpole M, Collen B et al (2010) Global biodiversity: indicators of recent declines. Science 328(5982):1164–1168. https://doi.org/10.1126/science.1187512
Bylemans J, Gleeson DM, Lintermans M et al (2018) Monitoring riverine fish communities through eDNA metabarcoding: determining optimal sampling strategies along an altitudinal and biodiversity gradient. Metabarcoding Metagenomics 2:1–12. https://doi.org/10.3897/mbmg.2.30457
Cantera I, Cilleros K, Valentini A et al (2019) Optimizing environmental DNA sampling effort for fish inventories in tropical streams and rivers. Sci Rep 9:3085. https://doi.org/10.1038/s41598-019-39399-5
Carraro L, Hartikainen H, Jokela J et al (2018) Estimating species distribution and abundance in river networks using environmental DNA. Proc Natl Acad Sci 115(46):11724–11729. https://doi.org/10.1073/pnas.1813843115
Deiner K, Altermatt F (2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9(2):e88786. https://doi.org/10.1371/journal.pone.0088786
Deiner K, Bik HM, Mächler E et al (2017) Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol Ecol 26:5872–5895. https://doi.org/10.1111/mec.14350
Doi H, Fukaya K, Oka S et al (2019) Evaluation of detection probabilities at the water-filtering and initial PCR steps in environmental DNA metabarcoding using a multispecies site occupancy model. Sci Rep 9:3581. https://doi.org/10.1038/s41598-019-40233-1
Dudgeon D, Arthington AH, Gessner MO et al (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biol Rev Camb Philos Soc 81:163–182. https://doi.org/10.1017/S1464793105006950
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Eichmiller JJ, Best E, Sorensen PW et al (2016) Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ Sci Technol 50:1859–1867. https://doi.org/10.1021/acs.est.5b05672
Evans NT, Li Y, Renshaw MA et al (2017) Fish community assessment with eDNA metabarcoding: effects of sampling design and bioinformatic filtering. Can J Fish Aquat Sci 1374:1–13. https://doi.org/10.1139/cjfas-2016-0306
Ficetola GF, Taberlet P, Coissac E (2016) How to limit false positives in environmental DNA and metabarcoding? Mol Ecol Resour 16:604–607. https://doi.org/10.1111/1755-0998.12508
Guillera-Arroita G, Lahoz-Monfort JJ, van Rooyen AR et al (2017) Dealing with false-positive and false-negative errors about species occurrence at multiple levels. Methods Ecol Evol 8:1081–1091. https://doi.org/10.1111/2041-210X.12743
Hänfling B, Lawson Handley L, Read DS et al (2016) Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol 25(13):3101–3119. https://doi.org/10.1111/mec.13660
Hayami K, Sakata MK, Inagawa T et al (2020) Effects of sampling seasons and locations on fish environmental DNA metabarcoding in dam reservoirs. Ecol Evol 10(12):5354–5367. https://doi.org/10.1002/ece3.6279
Jacobsen CS, Nielsen TK, Vester JK et al (2018) Inter-laboratory testing of the effect of DNA blocking reagent G2 on DNA extraction from low-biomass clay samples. Sci Rep 8:5711. https://doi.org/10.1038/s41598-018-24082-y
Jane SF, Wilcox TM, Mckelvey KS et al (2015) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15:216–227. https://doi.org/10.1111/1755-0998.12285
Japanese Industrial Standards Committee (2009) Test method for particle size distribution of soils (JIS A1204)
Jerde CL, Olds BP, Shogren AJ et al (2016) Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environ Sci Technol 50:8770–8779. https://doi.org/10.1021/acs.est.6b01761
Kanbar HJ, Olajos F, Englund G, Holmboe M (2020) Geochemical identification of potential DNA-hotspots and DNA-infrared fingerprints in lake sediments. Appl Geochem. https://doi.org/10.1016/j.scitotenv.2019.135577
Kessler EJ, Davis MA, Ash KT et al (2019) Radiotelemetry reveals effects of upstream biomass and UV exposure on environmental DNA occupancy and detection for a large freshwater turtle. Environ DNA 2(1):13–23. https://doi.org/10.1002/edn3.42
Kimura K, Saitou K, Moriue Y (2015) Third report of freshwater fish fauna in Chigasaki City. Bull Chigasaki City Museum Heritage 24:21–46.
Kouduka M, Suko T, Morono Y et al (2012) A new DNA extraction method by controlled alkaline treatments from consolidated subsurface sediments. FEMS Microbiol Lett 326:47–54. https://doi.org/10.1111/j.1574-6968.2011.02437.x
Mächler E, Deiner K, Spahn F, Altermatt F (2016) Fishing in the water: effect of sampled water volume on environmental DNA-based detection of macroinvertebrates. Environ Sci Technol 50:305–312. https://doi.org/10.1021/acs.est.5b04188
Minamoto T, Hayami K, Sakata MK, Imamura A (2019) Real-time polymerase chain reaction assays for environmental DNA detection of three salmonid fish in Hokkaido, Japan: application to winter surveys. Ecol Res 34:237–242. https://doi.org/10.1111/1440-1703.1018
Miya M, Sato Y, Fukunaga T et al (2015) MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci 2:150088. https://doi.org/10.1098/rsos.150088
Miya M, Minamoto T, Yamanaka H et al (2016) Use of a filter cartridge for filtration of water samples and extraction of environmental DNA. J Vis Exp 117:e54741. https://doi.org/10.3791/54741
Nakagawa H, Yamamoto S, Sato Y et al (2018) Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw Biol 63:569–580. https://doi.org/10.1111/fwb.13094
Nukazawa K, Hamasuna Y, Suzuki Y (2018) Simulating the advection and degradation of the environmental DNA of common carp along a river. Environ Sci Technol 52:10562–10570. https://doi.org/10.1021/acs.est.8b02293
Oka S, Doi H, Miyamoto K et al (2020) Environmental DNA metabarcoding for biodiversity monitoring of a highly diverse tropical fish community in a coral reef lagoon: Estimation of species richness and detection of habitat segregation. Environ DNA. https://doi.org/10.1002/edn3.132
R Core Team (2019). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved from https://www.R-project.org/
Sakata MK, Maki N, Sugiyama H, Minamoto T (2017) Identifying a breeding habitat of a critically endangered fish, Acheilognathus typus, in a natural river in Japan. Sci Nat 104:100. https://doi.org/10.1007/s00114-017-1521-1
Sakata MK, Yamamoto S, Gotoh RO et al (2020) Sedimentary eDNA provides different information on timescale and fish species composition compared with aqueous eDNA. Environ DNA 2(4):505–518 https://doi.org/10.1002/edn3.75
Sato H, Sogo Y, Doi H, Yamanaka H (2017) Usefulness and limitations of sample pooling for environmental DNA metabarcoding of freshwater fish communities. Sci Rep 7:14860. https://doi.org/10.1038/s41598-017-14978-6
Schultz MT, Lance RF (2015) Modeling the sensitivity of field surveys for detection of environmental DNA (eDNA). PLoS ONE 10(10):e0141503. https://doi.org/10.1371/journal.pone.0141503
Shaw JLA, Clarke LJ, Wedderburn SD et al (2016) Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol Conserv 197:131–138. https://doi.org/10.1016/j.biocon.2016.03.010
Shogren AJ, Tank JL, Andruszkiewicz E et al (2017) Controls on eDNA movement in streams: transport, retention, and resuspension /704/158/2464 /704/242 /45/77 article. Sci Rep 7:5065. https://doi.org/10.1038/s41598-017-05223-1
Siegenthaler A, Wangensteen OS, Soto AZ et al (2019) Metabarcoding of shrimp stomach content: harnessing a natural sampler for fish biodiversity monitoring. Mol Ecol Resour 19:206–220. https://doi.org/10.1111/1755-0998.12956
Sigsgaard EE, Torquato F, Frøslev TG et al (2019) Using vertebrate environmental DNA from seawater in biomonitoring of marine habitats. Conserv Biol 34(3):697–710. https://doi.org/10.1111/cobi.13437
Strickler KM, Fremier AK, Goldberg CS (2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol Conserv 183:85–92. https://doi.org/10.1016/j.biocon.2014.11.038
The eDNA Society (2019) Environmental DNA sampling and experiment manual. Version 2.1. https://ednasociety.org/eDNA_manual_Eng_v2_1_3b.pdf
Thomsen PF, Kielgast J, Iversen LL et al (2012) Monitoring endangered freshwater biodiversity using environmental DNA. Mol Ecol 21:2565–2573. https://doi.org/10.1111/j.1365-294X.2011.05418.x
Turner CR, Uy KL, Everhart RC (2015) Environmental DNA fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol Conserv 183:93–102. https://doi.org/10.1016/j.biocon.2014.11.017
Valentini A, Taberlet P, Miaud C et al (2016) Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol Ecol 25:929–942. https://doi.org/10.1111/mec.13428
Wilcox TM, McKelvey KS, Young MK et al (2016) Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol Conserv 194:209–216. https://doi.org/10.1016/j.biocon.2015.12.023
WWF (2018) Living planet report—2018: aiming higher. https://www.worldwildlife.org/pages/living-planet-report-2018
Yamamoto S, Masuda R, Sato Y et al (2017) Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci Rep 7:40368. https://doi.org/10.1038/srep40368
Yamanaka H, Minamoto T, Matsuura J et al (2017) A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant. Limnology 18:233–241. https://doi.org/10.1007/s10201-016-0508-5
Acknowledgements
We would like to thank Mr. R. Osawa (Kobe University) for assistance with filtration. This study was partly supported by the Japan Society for the Promotion of Science (JSPS; KAKENHI Grant Numbers 19J11126 and 20H03326) and by a fund from a private company to which five of the authors (T.W., N.M., K.I., T.K., and H.O.) belong.
Author information
Authors and Affiliations
Contributions
MKS, TW, and TM conceived and designed the research. NM, KI, TK, and HO collected samples and performed filtration. MKS, HY, TS, and MM performed the experiments, along with bioinformatic and statistical analyses. MKS wrote the first draft of the manuscript. All authors discussed the results and contributed to the development of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
T.W., N.M., K.I., T.K., and H.O. belong to a private company.
Research involving animal rights
No animal experiments were performed in this study. All experiments were performed according to the current laws of Japan.
Additional information
Handling Editor: Teruhiko Takahara.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sakata, M.K., Watanabe, T., Maki, N. et al. Determining an effective sampling method for eDNA metabarcoding: a case study for fish biodiversity monitoring in a small, natural river. Limnology 22, 221–235 (2021). https://doi.org/10.1007/s10201-020-00645-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10201-020-00645-9