Abstract
Background and purpose
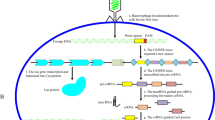
The central nervous system (CNS) faces unique difficulties in attaining permanent therapy for neurodegenerative disorder (ND). Genomic level forms of therapy have garnered interest in the recent decade, with the novel CRISPR/Cas9 gene editing tool continuing to be explored due to its efficiency, safety, and adaptability to varying conditions. With the aid of viral vectors as transport vectors, the gene editing tool has produced promising in vitro and in vivo findings in study models. Thus, this review focuses on the recent advancements and update of CRISPR/Cas9 to combat neurodegenerative diseases.
Methods
Articles detailing potential applications of CRISPR/Cas9 in neurodegenerative settings were retrieved from PubMed and Google Scholar with the keywords “CRISPR,” “gene editing,” and “neurodegenerative diseases.” Relevant information was collected and critically reviewed.
Results
The utility of CRISPR/Cas9 coupled with viral transduction ranges from the disruption of amyloid precursor protein (APP) production at a genomic level in Alzheimer’s disease (AD) to the deletion of varying exon portions of the Dmd gene in Duchenne muscular dystrophy (DMD) which would increase dystrophin expression. This usage of CRISPR/Cas9 also extends to experimentally ameliorate the neurodegenerative effects caused by viral infections.
Conclusion
The CRISPR/Cas9 gene editing tool is a powerful arsenal in the field of gene therapy and molecular medicine; hence, more research should be called to focus on the ample potential this tool has to offer in the field of neurodegenerative diseases.
Similar content being viewed by others
References
Nathan PJ, Cobb SR, Lu B, Bullmore ET, Davies CH (2011) Studying synaptic plasticity in the human brain and opportunities for drug discovery. Curr Opin Pharmacol 11:540–548. https://doi.org/10.1016/j.coph.2011.06.008
Bozyczko-Coyne D, Williams M (2007) Neurodegeneration. Compr Med Chem II, Elsevier p. 193–228. https://doi.org/10.1016/B0-08-045044-X/00168-1
Veazey C, Ozlem Erden Aki S, Cook KF, Lai EC, Kunik ME (2005) Prevalence and treatment of depression in Parkinson’s disease. J Neuropsychiatr Clin Neurosci 17:310–323. https://doi.org/10.1176/jnp.17.3.310
Sasmita AO, Kuruvilla J, Ling APK (2018) Harnessing neuroplasticity: modern approaches and clinical future. Int J Neurosci: 01–40. https://doi.org/10.1080/00207454.2018.1466781
Bankiewicz KS, Eberling JL, Kohutnicka M, Jagust W, Pivirotto P, Bringas J, Cunningham J, Budinger TF, Harvey-White J (2000) Convection-enhanced delivery of AAV vector in parkinsonian monkeys; in vivo detection of gene expression and restoration of dopaminergic function using pro-drug approach. Exp Neurol 164:2–14. https://doi.org/10.1006/exnr.2000.7408
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
Nayerossadat N, Ali P, Maedeh T (2012) Viral and nonviral delivery systems for gene delivery. Adv Biomed Res 1:27. https://doi.org/10.4103/2277-9175.98152
Bouard D, Alazard-Dany N, Cosset F-L (2009) Viral vectors: from virology to transgene expression. Br J Pharmacol 157:153–165. https://doi.org/10.1038/bjp.2008.349
Holehonnur R, Lella SK, Ho A, Luong JA, Ploski JE (2015) The production of viral vectors designed to express large and difficult to express transgenes within neurons. Mol Brain 8:12. https://doi.org/10.1186/s13041-015-0100-7
Abordo-Adesida E, Follenzi A, Barcia C, Sciascia S, Castro MG, Naldini L, Lowenstein PR (2005) Stability of lentiviral vector-mediated transgene expression in the brain in the presence of systemic antivector immune responses. Hum Gene Ther 16:741–751. https://doi.org/10.1089/hum.2005.16.741
Sengupta R, Mukherjee C, Sarkar N, Sun Z, Lesnik J, Huang J, Lu B (2016) An optimized protocol for packaging pseudotyped integrase defective lentivirus. Biol Proced Online 18:14. https://doi.org/10.1186/s12575-016-0044-z
Schambach A, Zychlinski D, Ehrnstroem B, Baum C (2013) Biosafety features of lentiviral vectors. Hum Gene Ther 24:132–142. https://doi.org/10.1089/hum.2012.229
Wanisch K, Yáñez-Muñoz RJ (2009) Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther 17:1316–1332. https://doi.org/10.1038/mt.2009.122
Chen X, Gonçalves MAFV (2016) Engineered viruses as genome editing devices. Mol Ther 24:447–457. https://doi.org/10.1038/mt.2015.164
Lerchner W, Corgiat B, Der Minassian V, Saunders RC, Richmond BJ (2014) Injection parameters and virus dependent choice of promoters to improve neuron targeting in the nonhuman primate brain. Gene Ther 21:233–241. https://doi.org/10.1038/gt.2013.75
Asokan A, Schaffer DV, Jude Samulski R (2012) The AAV vector toolkit: poised at the clinical crossroads. Mol Ther 20:699–708. https://doi.org/10.1038/mt.2011.287
Ai J, Li J, Gessler DJ, Su Q, Wei Q, Li H, Gao G (2017) Adeno-associated virus serotype rh.10 displays strong muscle tropism following intraperitoneal delivery. Sci Rep 7:40336. https://doi.org/10.1038/srep40336
Xie J, Xie Q, Zhang H, Ameres SL, Hung J-H, Su Q, He R, Mu X, Seher Ahmed S, Park S, Kato H, Li C, Mueller C, Mello CC, Weng Z, Flotte TR, Zamore PD, Gao G (2011) MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther 19:526–535. https://doi.org/10.1038/mt.2010.279
Vagner T, Dvorzhak A, Wójtowicz AM, Harms C, Grantyn R (2016) Systemic application of AAV vectors targeting GFAP-expressing astrocytes in Z-Q175-KI Huntington’s disease mice. Mol Cell Neurosci 77:76–86. https://doi.org/10.1016/j.mcn.2016.10.007
Rosenberg JB, Sondhi D, Rubin DG, Monette S, Chen A, Cram S, de BP, Kaminsky SM, Sevin C, Aubourg P, Crystal RG (2014) Comparative efficacy and safety of multiple routes of direct CNS administration of adeno-associated virus gene transfer vector serotype rh.10 expressing the human arylsulfatase a cDNA to nonhuman primates. Hum Gene Ther Clin Dev 25:164–177. https://doi.org/10.1089/humc.2013.239
Kay MA, Glorioso JC, Naldini L (2001) Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med 7:33–40. https://doi.org/10.1038/83324
Yang B, Li S, Wang H, Guo Y, Gessler DJ, Cao C, Su Q, Kramer J, Zhong L, Ahmed SS, Zhang H, He R, Desrosiers RC, Brown R, Xu Z, Gao G (2014) Global CNS transduction of adult mice by intravenously delivered rAAVrh.8 and rAAVrh.10 and nonhuman primates by rAAVrh.10. Mol Ther 22:1299–1309. https://doi.org/10.1038/mt.2014.68
Murlidharan G, Sakamoto K, Rao L, Corriher T, Wang D, Gao G, Sullivan P, Asokan A (2016) CNS-restricted transduction and CRISPR/Cas9-mediated gene deletion with an engineered AAV vector. Mol Ther Nucleic Acids 5:e338. https://doi.org/10.1038/mtna.2016.49
Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F (2015) In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33:102–106. https://doi.org/10.1038/nbt.3055
Kim EJ, Kang KH, Ju JH (2017) CRISPR-Cas9: a promising tool for gene editing on induced pluripotent stem cells. Korean J Intern Med 32:42–61. https://doi.org/10.3904/kjim.2016.198
He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B (2016) Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res 44:e85–e85. https://doi.org/10.1093/nar/gkw064
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542. https://doi.org/10.1038/nbt.3190
Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L et al (2013) Optical control of mammalian endogenous transcription and epigenetic states. Nature 500:472–476. https://doi.org/10.1038/nature12466
Sweatt JD (2013) The emerging field of neuroepigenetics. Neuron 80:624–632. https://doi.org/10.1016/j.neuron.2013.10.023
Marks WJ, Bartus RT, Siffert J, Davis CS, Lozano A, Boulis N et al (2010) Gene delivery of AAV2-neurturin for Parkinson’s disease: a double-blind, randomised, controlled trial. Lancet Neurol 9:1164–1172. https://doi.org/10.1016/S1474-4422(10)70254-4
Perdomini M, Belbellaa B, Monassier L, Reutenauer L, Messaddeq N, Cartier N, Crystal RG, Aubourg P, Puccio H (2014) Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich’s ataxia. Nat Med 20:542–547. https://doi.org/10.1038/nm.3510
Chen Y, Xiong M, Dong Y, Haberman A, Cao J, Liu H, Zhou W, Zhang SC (2016) Chemical control of grafted human PSC-derived neurons in a mouse model of Parkinson’s disease. Cell Stem Cell 18:817–826. https://doi.org/10.1016/j.stem.2016.03.014
Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, Sun X, Qin Z, Jin P, Li S, Li XJ (2017) CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest 127:2719–2724. https://doi.org/10.1172/JCI92087
György B, Lööv C, Zaborowski MP, Takeda S, Kleinstiver BP, Commins C, Kastanenka K, Mu D, Volak A, Giedraitis V, Lannfelt L, Maguire CA, Joung JK, Hyman BT, Breakefield XO, Ingelsson M (2018) CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol Ther Nucleic Acids 11:429–440. https://doi.org/10.1016/j.omtn.2018.03.007
Monteys AM, Ebanks SA, Keiser MS, Davidson BL (2017) CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther 25:12–23. https://doi.org/10.1016/j.ymthe.2016.11.010
Gaj T, Ojala DS, Ekman FK, Byrne LC, Limsirichai P, Schaffer DV (2017) In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci Adv 3:eaar3952. https://doi.org/10.1126/sciadv.aar3952
Lin H, Hu H, Duan W, Liu Y, Tan G, Li Z, Liu YK, Deng BB, Song XQ, Wang W, Wen D, Wang Y, Li CY (2018) Intramuscular delivery of scAAV9-hIGF1 prolongs survival in the hSOD1G93A ALS mouse model via upregulation of D-amino acid oxidase. Mol Neurobiol 55:682–695. https://doi.org/10.1007/s12035-016-0335-z
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E et al (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351(80):400–403. https://doi.org/10.1126/science.aad5725
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC et al (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351(80):403–407. https://doi.org/10.1126/science.aad5143
Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX et al (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351(80):407–411. https://doi.org/10.1126/science.aad5177
Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS (2017) Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 8:14454. https://doi.org/10.1038/ncomms14454
Chou Y, Krupp A, Kaynor C, Gaudin R, Ma M, Cahir-McFarland E, Kirchhausen T (2016) Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Sci Rep 6:36921. https://doi.org/10.1038/srep36921
Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K (2015) CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS One 10:e0136046. https://doi.org/10.1371/journal.pone.0136046
Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K (2014) RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci 111:11461–11466. https://doi.org/10.1073/pnas.1405186111
Liao H-K, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Belmonte JCI (2015) Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6:6413. https://doi.org/10.1038/ncomms7413
van Diemen FR, Kruse EM, Hooykaas MJG, Bruggeling CE, Schürch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJHJ, Lebbink RJ (2016) CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog 12:e1005701. https://doi.org/10.1371/journal.ppat.1005701
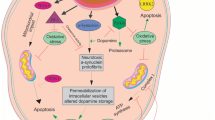
Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10:S10–S17. https://doi.org/10.1038/nm1066
Pires C, Schmid B, Petræus C, Poon A, Nimsanor N, Nielsen TT, Waldemar G, Hjermind LE, Nielsen JE, Hyttel P, Freude KK (2016) Generation of a gene-corrected isogenic control cell line from an Alzheimer’s disease patient iPSC line carrying a A79V mutation in PSEN1. Stem Cell Res 17:285–288. https://doi.org/10.1016/j.scr.2016.08.002
Ortiz-Virumbrales M, Moreno CL, Kruglikov I, Marazuela P, Sproul A, Jacob S, Zimmer M, Paull D, Zhang B, Schadt EE, Ehrlich ME, Tanzi RE, Arancio O, Noggle S, Gandy S (2017) CRISPR/Cas9-correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 N141I neurons. Acta Neuropathol Commun 5:77. https://doi.org/10.1186/s40478-017-0475-z
Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N (2011) Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet 20:4530–4539. https://doi.org/10.1093/hmg/ddr394
Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR III, Nakanishi N, Andreyev AY, Okamoto SI, Jaenisch R, Ambasudhan R, Lipton SA (2013) Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell 155:1351–1364. https://doi.org/10.1016/j.cell.2013.11.009
Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA et al (2012) Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74:1031–1044. https://doi.org/10.1016/j.neuron.2012.05.009
Yang S-H, Cheng P-H, Banta H, Piotrowska-Nitsche K, Yang J-J, Cheng ECH, Snyder B, Larkin K, Liu J, Orkin J, Fang ZH, Smith Y, Bachevalier J, Zola SM, Li SH, Li XJ, Chan AWS (2008) Towards a transgenic model of Huntington’s disease in a non-human primate. Nature 453:921–924. https://doi.org/10.1038/nature06975
Fitzner D, Simons M (2010) Chronic progressive multiple sclerosis – pathogenesis of neurodegeneration and therapeutic strategies. Curr Neuropharmacol 8:305–315. https://doi.org/10.2174/157015910792246218
Wokke J (1996) Riluzole. Lancet 348:795–799. https://doi.org/10.1016/S0140-6736(96)03181-9
Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA (2015) Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 6:6244. https://doi.org/10.1038/ncomms7244
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(80):819–823. https://doi.org/10.1126/science.1231143
Shishido-Hara Y (2015) Progressive multifocal leukoencephalopathy: dot-shaped inclusions and virus-host interactions. Neuropathology 35:487–496. https://doi.org/10.1111/neup.12203
Wang T, Rumbaugh JA, Nath A (2006) Viruses and the brain: from inflammation to dementia. Clin Sci 110:393–407. https://doi.org/10.1042/CS20050278
Boban J, Kozic D, Turkulov V, Ostojic J, Semnic R, Lendak D, Brkic S (2017) HIV-associated neurodegeneration and neuroimmunity: multivoxel MR spectroscopy study in drug-naïve and treated patients. Eur Radiol 27:4218–4236. https://doi.org/10.1007/s00330-017-4772-5
Ebina H, Misawa N, Kanemura Y, Koyanagi Y (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3:2510. https://doi.org/10.1038/srep02510
Packer RJ, Gurney JG, Punyko JA, Donaldson SS, Inskip PD, Stovall M, Yasui Y, Mertens AC, Sklar CA, Nicholson HS, Zeltzer LK, Neglia JP, Robison LL (2003) Long-term neurologic and neurosensory sequelae in adult survivors of a childhood brain tumor: childhood cancer survivor study. J Clin Oncol 21:3255–3261. https://doi.org/10.1200/JCO.2003.01.202
Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW et al (2017) Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 377:1713–1722. https://doi.org/10.1056/NEJMoa1706198
Coutelle C, Rodeck C (2002) On the scientific and ethical issues of fetal somatic gene therapy. Gene Ther 9:670–673. https://doi.org/10.1038/sj.gt.3301761
Ojala DS, Amara DP, Schaffer DV (2015) Adeno-associated virus vectors and neurological gene therapy. Neuroscience 21:84–98. https://doi.org/10.1177/1073858414521870
Jaenisch R, Zhang F, Gage F (2017) Genome editing in neurosciences. Springer International Publishing, Cham. https://doi.org/10.1007/978-3-319-60192-2
Laycock-van Spyk S, Thomas N, Cooper DN, Upadhyaya M (2011) Neurofibromatosis type 1-associated tumours: their somatic mutational spectrum and pathogenesis. Hum Genomics 5:623–690. https://doi.org/10.1186/1479-7364-5-6-623
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455. https://doi.org/10.1016/j.cell.2014.09.014
Huang J, Su Q, Yang J, Lv Y, He Y, Chen J et al (2015) Sample sizes in dosage investigational clinical trials: a systematic evaluation. Drug Des Devel Ther 9:305. https://doi.org/10.2147/DDDT.S76135
Meyer K, Ferraiuolo L, Schmelzer L, Braun L, McGovern V, Likhite S, Michels O, Govoni A, Fitzgerald J, Morales P, Foust KD, Mendell JR, Burghes AHM, Kaspar BK (2015) Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose–response study in mice and nonhuman primates. Mol Ther 23:477–487. https://doi.org/10.1038/mt.2014.210
Takahashi M, Suzuki M, Fukuoka M, Fujikake N, Watanabe S, Murata M, Wada K, Nagai Y, Hohjoh H (2015) Normalization of overexpressed α-synuclein causing Parkinson’s disease by a moderate gene silencing with RNA interference. Mol Ther Nucleic Acids 4:e241. https://doi.org/10.1038/mtna.2015.14
Aronin N, DiFiglia M (2014) Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov Disord 29:1455–1461. https://doi.org/10.1002/mds.26020
Long C, Amoasii L, Bassel-Duby R, Olson EN (2016) Genome editing of monogenic neuromuscular diseases. JAMA Neurol 73:1349–1355. https://doi.org/10.1001/jamaneurol.2016.3388
Baylis F, McLeod M (2018) First-in-human phase 1 CRISPR gene editing cancer trials: are we ready? Curr Gene Ther 17:309–319. https://doi.org/10.2174/1566523217666171121165935
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kuruvilla, J., Sasmita, A.O. & Ling, A.P.K. Therapeutic potential of combined viral transduction and CRISPR/Cas9 gene editing in treating neurodegenerative diseases. Neurol Sci 39, 1827–1835 (2018). https://doi.org/10.1007/s10072-018-3521-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-018-3521-0