Abstract
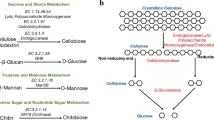
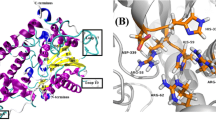
PACS and mathematical subject classification numbers as needed. Molecular dynamic simulation is a very usable tool to understand various factors, including structure temperature dependence, dynamics, and stability for protein structure. The three main components, namely endoglucanase, exoglucanase, and β-glucosidase, effectively convert lignocellulosic biomass into fermentable sugar. Cellulose is the major component of plant cell walls and is the most abundant organic compound on the earth. Somewhat organisms can use cellulose as a food source, possessing cellulases (cellobiohydrolases and endoglucanases) that can catalyze the hydrolysis of the β-(1,4) glycosidic bonds. In this work, we investigated conformational and structural properties of PcCel45A protein by changing at temperatures with 300 K, 333 K, and 352 K. We found that the ASN92 residue was the major contributor to the stability of structure; some other residues correlated significantly with thermal stability. We also compared the theoretical results of the current study with the experimental ones published in previous studies.






Similar content being viewed by others
References
Rosegrant MW, Zhu T, Msangi S, Sulser T (2008) Global scenarios for biofuels: impacts and implications. Rev Agri Econ 30:495–505
EUR-Lex - 32009L0028 - EN - EUR-Lex. https://eur-lex.europa.eu/eli/dir/2009/28/oj. Accessed 24 Jul 2019
Global biofuel production 2018. In: Statista. https://bit.ly/2LFl4w7. Accessed 24 Jul 2019
Biodiesel production United States 2018. In: Statista. https://bit.ly/30MWFrQ
Srivastava N, Srivastava M, Mishra PK, et al. (2015) Application of cellulases in biofuels industries: an overview. J Biof nd Bioen 1:55. https://doi.org/10.5958/2454-8618.2015.00007.3
Igarashi K, Ishida T, Hori C, Samejima M (2008) Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete phanerochaete chrysosporium. Appl Environ Microbiol 74:5628–5634. https://doi.org/10.1128/AEM.00812-08
Kricka W, Fitzpatrick J, Bond U (2015) Challenges for the production of bioethanol from biomass using recombinant yeasts. In: Advances in applied microbiology. Elsevier, pp 89–125
Eriksson K-EL, Blanchette RA, Ander P (1990) Biodegradation of cellulose. In: Microbial and enzymatic degradation of wood and wood components. Springer, Berlin
Clarke AJ (1997) Biodegradation of cellulose: enzymology and biotechnology. Technomic Pub Co, Lancaster
Rojas OJ, Jeong C, Turon X (2007) American chemical measurement of cellulase activity with piezoelectric resonators. In: Argyropoulos DS (ed) Materials, chemicals, and energy from forest biomass. American Chemical Society, Washington, DC, pp 478–494
Bhat MK, Bhat S (1997) Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv 15:583–620. https://doi.org/10.1016/S0734-9750(97)00006-2
Shewmaker CK, Stalker DM (1992) Modifying starch biosynthesis with transgenes in potatoes. Plant Physiol 100:1083–1086. https://doi.org/10.1104/pp.100.3.1083
Müller-Röber B, Sonnewald U, Willmitzer L (1992) Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J 11:1229–1238
Oakes JV, Shewmaker CK, Stalker DM (1991) Production of cyclodextrins, a novel carbohydrate, in the tubers of transgenic potato plants. Biotechnology (NY) 9:982–986
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43. https://doi.org/10.1128/MMBR.65.1.1-43.2001
Sauer DB, Karpowich NK, Song JM, Wang D -N (2015) Rapid bioinformatic identification of thermostabilizing mutations. Biophys J 109:1420–1428
Pucci F, Rooman M (2017) Physical and molecular bases of protein thermal stability and cold adaptation. Curr Opin Struct Biol 42:117–128
Yennamalli RM, Rader AJ, Kenny AJ, et al. (2013) Endoglucanases: insights into thermostability for biofuel applications. Biotechnol Biofuels 6:136
Karlsson J, Siika-aho M, Tenkanen M, Tjerneld F (2002) Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J Biotechnol 99:63–78
Karlsson J, Siika-aho M, Tenkanen M, Tjerneld F (2002) Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J Biotechnol 99:63–78. https://doi.org/10.1016/S0168-1656(02)00156-6
Godoy AS, Ramia MP, Camilo CM, Polikarpov I X-ray structure of PcCel45A expressed in Aspergillus nidullans. To be published. https://doi.org/10.2210/pdb5kjo/pdb
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105:6474–6487. https://doi.org/10.1021/jp003919d
Abraham MJ, Murtola T, Schulz R, et al. (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Páll S, Abraham MJ, Kutzner C, et al. (2015) Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In: Markidis S, Laure E (eds) Solving software challenges for exascale. Springer International Publishing, Cham
Pronk S, Páll S, Schulz R, et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. https://doi.org/10.1093/bioinformatics/btt055
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. https://doi.org/10.1021/ct700301q
Van Der Spoel D, Lindahl E, Hess B, et al. (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Jorgensen WL, Chandrasekhar J, Madura JD, et al. (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. https://doi.org/10.1063/1.445869
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101. https://doi.org/10.1063/1.2408420
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log(N ) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Pettersen EF, Goddard TD, Huang CC, et al. (2004) UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Peng X-N, Wang J, Zhang W (2017) Molecular dynamics simulation analysis of the effect of T790M mutation on epidermal growth factor receptor protein architecture in non-small cell lung carcinoma. Oncol Lett 14:2249–2253. https://doi.org/10.3892/ol.2017.6387
Nishiyama K (2008) Thermal behavior of luciferase on nanofabricated hydrophilic Si surface. Biomacromolecules 9:1081–1083. https://doi.org/10.1021/bm701264r
Martinez R, Schwaneberg U, Roccatano D (2011) Temperature effects on structure and dynamics of the psychrophilic protease subtilisin S41 and its thermostable mutants in solution. Protein Eng Des Selection 24:533–544
Nakamura A, Ishida T, Kusaka K et al (2015) Newton’s cradle proton relay with amide–imidic acid tautomerization in inverting cellulase visualized by neutron crystallography. Sci Adv 1:e1500263. https://doi.org/10.1126/sciadv.1500263
Godoy AS, Pereira CS, Ramia MP, et al. (2018) Structure, computational and biochemical analysis of PcCel45A endoglucanase from Phanerochaete chrysosporium and catalytic mechanisms of GH45 subfamily C members. Sci Rep 8:3678
Szijártó N, Siika-aho M, Tenkanen M, et al. (2008) Hydrolysis of amorphous and crystalline cellulose by heterologously produced cellulases of Melanocarpus albomyces. J Biotechnol 136:140–147. https://doi.org/10.1016/j.jbiotec.2008.05.010
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Unal, M.A., Boyacioglu, B., Unver, H. et al. Molecular simulation of PcCel45A protein expressed from Aspergillus nidulans to understand its structure, dynamics, and thermostability. J Mol Model 25, 317 (2019). https://doi.org/10.1007/s00894-019-4175-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-019-4175-4