Abstract
Objectives
Crohn’s disease patients, who are prone to develop periodontal diseases, may carry genetic defects in their Th17 cytokine, human beta-defensin (hBD) 1–3, and salivary and scavenger agglutinin (SALSA) expressions. Biochemical composition of saliva reflects the oral consequences of systemic immune response modifications. Our aim was to evaluate the salivary Th17 cytokine, epithelial hBD 1–3, and SALSA levels in relation to Crohn’s disease.
Materials and methods
This cross-sectional study included 42 Crohn’s disease patients and 34 systemically healthy controls. Periodontal and dental indexes were measured, and stimulated saliva samples were collected. Salivary Th17 cytokine levels were analyzed by multiplex technique, and hBD 1–3 and SALSA levels by enzyme-linked immunosorbent assay.
Results
There were 19 gingivitis and 11 initial periodontitis patients in the Crohn’s disease group, and 15 gingivitis and 4 initial periodontitis in the control group. In comparison to controls, higher salivary Th17 cytokine levels were observed in Crohn’s disease patients. No statistical difference was observed between Crohn’s disease and control groups in terms of their salivary hBD 1–3 and SALSA levels. Based on the regression analysis, there is no independent association between Crohn’s disease and salivary Th17 cytokine levels.
Conclusions
Crohn’s disease does not relate to salivary antimicrobial hBD 1–3 or SALSA levels. While Crohn’s disease patients have higher salivary Th17 cytokine levels in comparison to systemically healthy controls, an independent association between Crohn’s disease and Th17 cytokine profile is still missing.
Clinical relevance
Diminished Th17 cytokine response in Crohn’s disease, which might be related to genetic susceptibility, can be also visualized in saliva.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crohn’s disease is a type of inflammatory bowel disease characterized by the chronic transmural intestinal inflammation. It can affect any part of the gastrointestinal track from the oral cavity to the anus [1]. It has a relapsing–remitting character, and its prevalence varies between 3 and 20 in 100,000 individuals, being highest in the industrialized countries [2]. Major risk factors of the Crohn’s disease are environmental exposures (diet, life style, smoking, and medications), dyscolonization of microbiota, disrupted immune response, and genetic susceptibility. Genetic risk ratio of Crohn’s disease is generally higher than that of other complex diseases, including type 2 diabetes mellitus, schizophrenia, or celiac disease [3]. Single nucleotide polymorphism (SNP)-based genome wide association studies pointed out over 140 loci that are related to the Crohn’s disease [4]. Among the Crohn’s disease-associated genes, a strong and replicated association was observed in IL23R gene, which takes part in Th17 lymphocyte differentiation [5]. In turn, high levels of serum interleukin (IL)-6, IL-17A, IL-23, and interferon-γ are commonly observed Crohn’s disease patients, especially when they are at the active state of the disease [6]. Earlier studies also indicated that human beta-defensins (hBDs), which are chemotactic to T cells and induce the mRNA expression of IL-1β, IL-6, and IL-22, may have copy number variations in Crohn’s disease [7]. When tissue and circulating hBD protein levels were analyzed, elevated hBD-3 levels were detected in terminal ileum, and elevated hBD-2 levels were detected in serum samples of Crohn’s disease patients [8]. IL-22 overexpression exacerbates mucosal inflammation and upregulates the expression of the salivary scavenger and agglutinin (SALSA) protein, which is also known as salivary agglutinin SAG, gp340, or DMBT1 [9]. SALSA is an innate immune protein found on mucosal surfaces and in secretions. It interacts with variety of microbial and host ligands, and appears to support immunological homeostasis [9,10,11]. Indeed, a deletion variant of DMBT1 with a reduced number of scavenger receptor cysteine-rich domain coding exons is associated with increased risk of Crohn’s disease [12], and diminished SALSA function has been associated with Crohn’s disease [9].
Periodontal diseases and caries are infection-induced diseases of the oral cavity. While dysbiotic biofilms are the prerequisite for the onset of these diseases, systemic and environmental conditions that dysregulate host immune responses contribute to the disease pathogenesis as well [13]. Crohn’s disease patients usually suffer from oral infectious diseases, including apthous-ulcerative lesions, caries, and periodontal diseases [14]. In fact, recent evidence suggests that Crohn’s disease is associated with severe periodontitis and early teeth-loss [15, 16]. On one hand, extension of gut mucosal inflammation was proposed as an explanatory factor for the high prevalence of periodontitis observed in Crohn’s disease [16]. On the other hand, both periodontitis and Crohn’s disease are multifactorial diseases and share similarities in their pathogenesis, including poor oral hygiene, smoking, diet, psychosocial stress, disturbed cytokine network or disrupted antimicrobial response [15, 17]. Indeed, previous studies suggested associations between periodontal inflammation and disrupted intraoral SALSA and hBD levels [18, 19]. Nevertheless, the observed changes on the oral immune response can arouse from the simultaneous impact of Crohn’s disease and oral inflammatory diseases, which is not yet fully elucidated.
Saliva contains large variety of molecules that originate from oral cavity as well as serum-derived markers. Non-invasively collected saliva contains biological markers that are useful to diagnose periodontal diseases and caries, detect oral infection and inflammation in a non-dental setting, and present information about the overall health of the patient [20]. Two common infectious diseases of the oral cavity, periodontitis and caries, can regulate the levels of various salivary biomarkers either by stimulating immune cell responses, or by affecting the oral environment (pH, oxidative stress) or by modulating the salivary microbial composition [21, 22]. Our group recently demonstrated the independent associations between Crohn’s disease and salivary immunoglobulin (Ig) antibody responses against two periodontitis-associated bacteria, Porphyromonas gingivalis and Prevotella intermedia [23]. In the present study, we hypothesized that the genetic background of Crohn’s disease causes defects in oral immune and antimicrobial protein responses. Thus, we analyzed the salivary Th17 cytokine, epithelial hBD 1–3, and SALSA levels in relation to Crohn’s disease. As the salivary antimicrobial response protein levels can be affected by periodontitis [21] and caries [22], regression models were applied to demonstrate independent associations between Crohn’s disease and salivary Th17 cytokine, epithelial hBD 1–3, and SALSA levels.
Materials and methods
Patient recruitment
The patient recruitment, clinical evaluations, and saliva collection of this case–control study were performed between January 2017 and May 2018. The recruitment of Crohn’s disease patients was performed by publishing an invitation advertisement in the patient journal of Crohn and Colitis patient organization (IBD Association of Finland). For all patients, the diagnosis of Crohn’s disease was confirmed by endoscopy and histopathological analysis of intestinal biopsy specimens were prerequisite. To recruit the control group without Crohn’s disease or other immune system disorders, advertisements placed at the Institute of Dentistry, University of Turku.
Exclusion criteria for both groups were as follows: being diagnosed with diabetes mellitus or periodontitis as a manifestation of systemic disease [24], being diagnosed with moderate to severe periodontitis, being smoker, pregnant or lactating, excessive use of alcohol, having less than 24 teeth, and use of antimicrobial medication in the preceding 6 months prior to the clinical examination.
As a result, 42 Crohn’s disease patients and 34 systemically healthy individuals, who were willing to participate and fit to the above mentioned inclusion criteria, were recruited.
Clinical oral examinations and saliva collection
Oral mucosal examinations of all participants were performed by a specialist in oral pathology (JR). All pathologic findings were registered describing the appearance and location of the lesion. Clinical photographs were taken when necessary.
Fiber-optic transillumination and tactile examination were used during the evaluation of cariological status. Caries lesions, erosion, and attrition were registered. Caries prevalence was described using the decayed-missed-filled teeth (DMFT) index.
A WHO probe (LM-Instruments Oy, Parainen, Finland) with a ball-tip end diameter of 0.5 mm was used in evaluation of periodontal status. Visible plaque index, bleeding on probing, gingival recession, probing pocket depth, and clinical attachment level measurements were performed at six sites per each natural tooth and dental implant. In addition, tooth mobility and furcation defects were recorded when present. The periodontal disease classifications of the patients were determined according to the 2018 Periodontal Disease Classification [25]. All dental and periodontal evaluations were performed by a calibrated periodontist (MG).
Collection of saliva samples
Participants were asked not to eat or brush their teeth 30 min prior to clinical examinations. Paraffin-stimulated saliva samples were collected from each participant. All appointments were during afternoon hours. Salivary flow rates were recorded. All saliva samples were aliquoted and stored as whole saliva (for cytokine and hBD analysis) or stored with 10 mM EDTA (for SALSA analysis), both at − 70 °C.
Determination of salivary Th17 cytokine, hBD 1–3, and SALSA levels
Saliva samples were centrifuged at 10,000 rpm for 5 min, and the supernatants were used to detect salivary cytokine and hBD levels. Salivary levels of IL-1β, IL-4, IL-6, IL-10, IL-17A, IL-17F, IL-21, IL-22, IL-23, IL-25, IL-31, IL-33, interferon (IFN)-γ, and soluble CD40L (sCD40L) were measured with the Luminex® xMAP™ technique (Luminex Corporation, Austin, TX, USA) using commercially available kits (Bio-Plex Pro™ Human Th17 Cytokine Panel, Bio-Rad, Santa Rosa, CA, USA). The lowest limit of detection (LOD) for each analyte was as follows: IL-1β, 0.1 pg/ml; IL-4, 1.9 pg/ml; IL-6, 2.0 pg/ml; IL-10, 1.4 pg/ml; IL-17A, 0.8 pg/ml; IL-17F, 3.1 pg/ml; IL-21, 2.5 pg/ml; IL-22, 0.9 pg/ml; IL-23, 5.2 pg/ml; IL-25, 0.8 pg/ml, IL-31, 2.1 pg/ml; IL-33, 1.6 pg/ml; IFN-γ, 0.3 pg/ml; and sCD40L, 4.4 pg/ml.
Salivary hBD-1, hBD-2, and hBD–3 were analyzed using commercial ELISA kits (Peprotech, catalog numbers 900-M202, 900-M172, 900-M210). The analyses were performed according to the manufacturer’s instructions. A 1:30 dilution was used for the hBD-1 analysis, and hBD-2 and hBD-3 were analyzed from undiluted samples. LOD levels for each hBD were as follows: 4 pg/ml for hBD-1, 16 pg/ml for hBD-2, and 62 pg/ml for hBD-3.
Salivary SALSA levels and protein level size variation were analyzed using commercial ELISA kit (Nordic Biosite, Täby, Sweden) and with western blot, respectively. For ELISA assays, the saliva samples were diluted 1:10 or 1:30 in dilution buffer provided with the kit. The analysis was performed according to manufacturer’s instructions. Western blot analysis was done as described earlier [13]. Briefly, saliva samples were separated by SDS–polyacrylamide gel electrophoresis and transferred to a PVDF membrane, 0.2-µm pore size (Bio-Rad). Membranes were blocked with 5% non-fat milk powder in TSB-T (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4) and stained with mAb 213–6 (dil 1:10 000, Bioporto diagnostics, Hellerup, Denmark) and HRP-conjugated anti-mouse antibody (1:10 000, Jackson ImmunoReseach Laboratories, West Grove, PA). The chemiluminescence of the membranes was documented with ChemiDoc MP imaging system (Bio-Rad), and the size of each protein was estimated using Image Lab™ molecular weight analysis tool (version 6.0.1, Bio-Rad Laboratories), and Precisions Plus protein standard (Bio-Rad) transferred to the membrane with the saliva samples.
Statistical analyses
All statistical analyses were performed using the SPSS statistical program (version 26.0; IBM Inc., Armonk, NY, USA). The one-way ANOVA test was used for comparing demographic and clinical data (with the exception of gender) between the Crohn’s disease and control groups. Gender distribution between the groups was compared with Fisher’s Exact test. Distribution Th17 cytokines, hBDs, and SALSA levels were skewed; therefore, Mann–Whitney U test was applied in data comparisons. Linear regression analyses were performed to analyze the associations between Crohn’s disease and the salivary analytes, after adjusting for age, DMFT, BOP%, and medication use. Statistical significance was defined as p < 0.05.
Results
Nineteen participants were diagnosed with gingivitis, 11 with initial periodontitis, and 12 with periodontal health in the Crohn’s disease group, and 15 participants with gingivitis, 4 with initial periodontitis, and 15 with periodontal health in the control group.
No significant difference was observed between the Crohn’s disease and control groups in terms of their age, gender, number of teeth, VPI%, BOP%, number of teeth with PPD ≥ 4 mm, and saliva flow rates. DMFT index scores, however, were significantly higher in Crohn’s disease group compared to the controls (p = 0.001, Table 1).
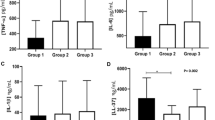
Elevated salivary IL-4, IL-6, IL-10, IL-17A, IL-17F, IL-22, IL-23, IL-25, IL-35, IFN-γ, and sCD40 levels were observed in Crohn’s disease group, in comparison to controls (Table 2). No difference in salivary levels of IL-1β, IL-31, hBD-1, hBD-2, hBD-3, or SALSA was observed between Crohn’s disease and control groups (Table 3).
In western blots, SALSA appears as slightly diffuse bands, due to its glycosylation. Yet, SALSA proteins with different sizes (270–317 kDa) of one or double bands, were evident in individual saliva samples (examples shown in Fig. 1), but no difference in size patterns was seen between groups.
Examples of different size variants of SALSA detected in saliva samples from Chron’s disease patients (A–C) and controls (D–F). Western blots of saliva samples were stained with anti-SALSA mAb, and different size of one or two slightly diffuse bands were detected. No difference was noted between the groups. Arrow indicates the 250 kDa standard
Linear regression models revealed significant associations between salivary IL-6 and IL-10 with Crohn’s disease; however, those associations were diminished after adjusting the model for age, DMFT, BOP%, and medication use (Table 4). Associations of BOP% and DMFT with salivary Th17 cytokine, hBD 1–3, and SALSA levels are presented in Table 5.
Discussion
To our knowledge, this study is the first to analyze salivary hBD-1, hBD-2, hBD-3, and SALSA levels in Crohn’s disease patients. According to our results, salivary hBD-1, hBD-2, hBD-3, and SALSA levels of Crohn’s disease patients are similar to those of systemically healthy controls, while salivary Th17 cytokine levels were found higher in Crohn’s disease patients. Nevertheless, according to the regression analysis, there is no independent association between elevated salivary Th17 cytokine levels and Crohn’s disease.
Comprehensive analysis of salivary Th17 cytokine, hBDs, and SALSA levels were the main strength of this study. Previous studies indicated that there are reciprocal interactions between the expressions of Th17 cytokine, hBD 1–3, and SALSA levels [7,8,9]. One limitation of this study was the relatively low participant numbers. Participants with Crohn’s disease were recruited from the Crohn and Colitis patient organization (IBD Association of Finland) between January 2017 and May 2018. All members of the association received the invitation and all who fit to the inclusion criteria were recruited. Therefore, no sample size calculation was performed. Secondly, the localization of the lesions and disease behavior according to the Montreal classification were not available; therefore, this information was not implemented into the current study. Thirdly, even though the periodontal disease diagnosis of participants was determined according to the 2018 Periodontal Disease Classification [25], use of WHO probe and the lack of radiographs restrained us to define the grades of periodontitis. As the maximum probing pocket depth measured from each periodontitis patient was 4 mm, these participants were diagnosed as initial periodontitis. Finally, majority of the Crohn’s disease patients were receiving anti-inflammatory or immunosuppressive drugs. While this was a standard procedure for this group of patients, it may still affect the cytokine and antimicrobial peptide expression profiles.
Th17 cytokine levels were previously evaluated in saliva, gingival crevicular fluid, dental biofilm, and gingival tissue samples of Crohn’s disease patients. Elevated gingival IL-10 RNA expression was documented in Crohn’s disease patients in relation to healthy controls, while no difference was observed for IL-2 and IL-6 [26]. When the gingival crevicular fluid Th17 cytokine protein expressions were analyzed in Crohn’s patients, similar expression profiles of IL-1β, IL-6, IL-10, IL-12p40, IL-12p70, IFN-γ, and tumor necrosis factor (TNF)-α were observed in Crohn’s disease patients and in systemically healthy controls [27]. According to our findings, salivary Th17 cytokine levels were elevated in Crohn’s disease patients; however, there are no independent associations between Crohn’s disease and elevated cytokine levels. Although a previous study showed higher salivary IL-1β and TNF-α levels in Crohn’s patients in comparison to systemically healthy controls, no information on the oral health status of the participants was given [28]. Indeed, similar to our findings, when the salivary samples were collected from individuals with healthy periodontium, no significant difference was observed in IL-1β, IL-10, IL-17A, and TNF-α levels between Crohn’s patients and healthy controls [29]. In inflammatory bowel disease patients, gingival and intestinal tissue Th17 cytokine expression profiles differ from each other [30], which indicates that the systemic condition is not the only determinant of oral cytokine expression, but the local environment, i.e., oral microbiome, also plays a significant role.
As mentioned above, to our knowledge, there is no available information on the oral hBD or SALSA levels of Crohn’s disease patients. hBDs are antimicrobial peptides with wide-range of functions, including the regulation of T cell chemotaxis, epithelial cell proliferation, and wound healing. In the oral cavity, hBDs are mainly secreted by epithelial cells [31]. Early studies associated Crohn’s disease with low copy number defects at the hBD-2 gene (DEFB4) region [7, 32]. Interestingly, studies by Fellermann et al., [7] and Bentley et al., [32] found the associations in opposite directions. However, neither genome wide association studies nor replication studies found evidence on the association between DEFB4 gene copy number variation and Crohn’s disease, indicating the false-positive results were possibly an outcome of limited statistical power and technological limitations [32, 33]. Similarly, an earlier study proposed association between Crohn’s disease and copy number defect at the gene DMBT1 [12] but this was not confirmed in a later study with other cohort [34]. In our study, we could see size variations of the salivary SALSA molecules between individuals, but no difference was observed between Crohn’s disease patients and control group. The noted size variation of SALSA may reflect to differences in protein core, but also the heavy glycosylation of SALSA affects the protein size and thus hampers the interpretation. Increased SALSA levels are reported in the intestine of Crohn’s disease patients [12], but based on our findings, salivary hBD-1, hBD-2, hBD-3, or SALSA levels did not differ between Crohn’s disease patients and systemically healthy controls. This is in line with the recent evidence that defects in the hBD or SALSA expressions do not necessarily exists in Crohn’s disease, as there is no genetic susceptibility. Yet, confounding factors, such as disease-specific quality of life, may significantly contribute to the dual interactions between Crohn’s disease and periodontal immune responses [35].
Conclusion
Our findings suggest that there is a tendency of increased salivary Th17 cytokine levels in Crohn’s disease patients; however, an independent association between Crohn’s disease and Th17 cytokine profile is lacking. The shifts in the oral cytokine levels of Crohn’s disease patients can be either an outcome of genetic background, or explained by the existence of periodontal diseases and mucosal lesions that are related to Crohn’s disease, or both.
References
Adolph TE, Meyer M, Schwärzler J, Mayr L, Grabherr F, Tilg H (2022) The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol 19:753–767. https://doi.org/10.1038/s41575-022-00658-y
Gajendran M, Loganathan P, Catinella AP, Hashash JG (2018) A comprehensive review and update on Crohn’s disease. Dis Mon 64:20–57. https://doi.org/10.1016/j.disamonth.2017.07.001
Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K (2006) Family and twin studies in inflammatory bowel disease. World J Gastroenterol 12:3668–3672. https://doi.org/10.3748/wjg.v12.i23.3668
Garza-Hernandez D, Estrada K, Trevino V (2022) Multivariate genome-wide association study models to improve prediction of Crohn’s disease risk and identification of potential novel variants. Comput Biol Med 145:105398. https://doi.org/10.1016/j.compbiomed.2022.105398
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH (2006) A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314:1461–1463. https://doi.org/10.1126/science.1135245
Ogawa K, Matsumoto T, Esaki M, Torisu T, Iida M (2012) Profiles of circulating cytokines in patients with Crohn’s disease under maintenance therapy with infliximab. J Crohns Colitis 6:529–535. https://doi.org/10.1016/j.crohns.2011.10.010
Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, Radlwimmer B, Stange EF (2006) A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet 79:439–448. https://doi.org/10.1086/505915
Meisch JP, Nishimura M, Vogel RM, Sung HC, Bednarchik BA, Ghosh SK, Fu P, McCormick T, Weinberg A, Levine AD (2013) Human β-defensin 3 peptide is increased and redistributed in Crohn’s ileitis. Inflamm Bowel Dis 19:942–953. https://doi.org/10.1097/MIB.0b013e318280b11a
Reichhardt MP, Holmskov U, Meri S (2017) SALSA-A dance on a slippery floor with changing partners. Mol Immunol 89:100–110. https://doi.org/10.1016/j.molimm.2017.05.029
Loimaranta V, Jakubovics NS, Hytönen J, Finne J, Jenkinson HF, Strömberg N (2005) Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun 73:2245–2252. https://doi.org/10.1128/IAI.73.4.2245-2252.2005
Polley S, Louzada S, Forni D, Sironi M, Balaskas T, Hains DS, Yang F, Hollox EJ (2015) Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc Natl Acad Sci U S A 112:5105–5110
Renner M, Bergmann G, Krebs I, End C, Lyer S, Hilberg F, Helmke B, Gassler N, Autschbach F, Bikker F, Strobel-Freidekind O, Gronert-Sum S, Benner A, Blaich S, Wittig R, Hudler M, Ligtenberg AJ, Madsen J, Holmskov U, Annese V, Latiano A, Schirmacher P, Amerongen AV, D’Amato M, Kioschis P, Hafner M, Poustka A, Mollenhauer J (2007) DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn’s disease. Gastroenterology 133:1499–1509. https://doi.org/10.1053/j.gastro.2007.08.007
Könönen E, Gursoy M, Gursoy UK (2019) Periodontitis: a multifaceted disease of tooth-supporting tissues. J Clin Med 8:1135. https://doi.org/10.3390/jcm8081135
Rautava J, Gürsoy UK, Kullström A, Könönen E, Sorsa T, Tervahartiala T, Gürsoy M (2020) An oral rinse active matrix metalloproteinase-8 point-of-care immunotest may be less accurate in patients with Crohn’s disease. Biomolecules 10:395. https://doi.org/10.3390/biom10030395
Bertl K, Burisch J, Pandis N, Bruckmann C, Klinge B, Stavropoulos A (2022) Periodontitis prevalence in patients with ulcerative colitis and Crohn’s disease - PPCC: a case-control study. J Clin Periodontol 49:1262–1274. https://doi.org/10.1111/jcpe.13615
Baima G, Muwalla M, Testa G, Mazza F, Bebars A, Perotto S, Vernero M, Massano A, Romano F, Ribaldone DG, Aimetti M (2023) Periodontitis prevalence and severity in inflammatory bowel disease: a case-control study. J Periodontol 94:313–322. https://doi.org/10.1002/JPER.22-0322
Keskin M, Zeidán-Chuliá F, Gursoy M, Könönen E, Rautava J, Gursoy UK (2015) Two cheers for Crohn’s disease and periodontitis: beta-defensin-2 as an actionable target to intervene on two clinically distinct diseases. OMICS 19:443–450. https://doi.org/10.1089/omi.2015.0077
Bostanci N, Selevsek N, Wolski W, Grossmann J, Bao K, Wahlander A, Trachsel C, Schlapbach R, Öztürk VÖ, Afacan B, Emingil G, Belibasakis GN (2018) Targeted proteomics guided by label-free quantitative proteome analysis in saliva reveal transition signatures from health to periodontal disease. Mol Cell Proteomics 17:1392–1409. https://doi.org/10.1074/mcp.RA118.000718
Özdemir M, Caglayan F, Bikker FJ, Pussinen P, Könönen E, Yamalik N, Gürsoy M, Fteita D, Nazmi K, Güncü GN, Pietiäinen M, Tolvanen M, Gürsoy UK (2020) Gingival tissue human beta-defensin levels in relation to infection and inflammation. J Clin Periodontol 47:309–318. https://doi.org/10.1111/jcpe.13227
Gürsoy UK, Kantarci A (2022) Molecular biomarker research in periodontology: a roadmap for translation of science to clinical assay validation. J Clin Periodontol 49:556–561. https://doi.org/10.1111/jcpe.13617
Sánchez-Medrano AG, Martinez-Martinez RE, Soria-Guerra R, Portales-Perez D, Bach H, Martinez-Gutierrez F (2023) A systematic review of the protein composition of whole saliva in subjects with healthy periodontium compared with chronic periodontitis. PLoS ONE 18:e0286079. https://doi.org/10.1371/journal.pone.0286079
Ahmad P, Hussain A, Carrasco-Labra A, Siqueira WL (2022) Salivary proteins as dental caries biomarkers: a systematic review. Caries Res 56:385–398. https://doi.org/10.1159/000526942
Gürsoy M, Rautava J, Pussinen P, Kristoffersen AK, Enersen M, Loimaranta V, Gürsoy UK (2023) Salivary IgA and IgG antibody responses against periodontitis-associated bacteria in Crohn’s disease. Int J Mol Sci 24:2385. https://doi.org/10.3390/ijms24032385
Jepsen S, Caton JG, Albandar JM, Bissada NF, Bouchard P, Cortellini P, Demirel K, de Sanctis M, Ercoli C, Fan J, Geurs NC, Hughes FJ, Jin L, Kantarci A, Lalla E, Madianos PN, Matthews D, McGuire MK, Mills MP, Preshaw PM, Reynolds MA, Sculean A, Susin C, West NX, Yamazaki K (2018) Periodontal manifestations of systemic diseases and developmental and acquired conditions: consensus report of workgroup 3 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Clin Periodontol 45(Suppl 20):S219–S229. https://doi.org/10.1111/jcpe.12951
Papapanou PN, Sanz M, Buduneli N, Dietrich T, Feres M, Fine DH, Flemmig TF, Garcia R, Giannobile WV, Graziani F, Greenwell H, Herrera D, Kao RT, Kebschull M, Kinane DF, Kirkwood KL, Kocher T, Kornman KS, Kumar PS, Loos BG, Machtei E, Meng H, Mombelli A, Needleman I, Offenbacher S, Seymour GJ, Teles R, Tonetti MS (2018) Periodontitis: consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Clin Periodontol 45(Suppl 20):S162–S170. https://doi.org/10.1111/jcpe.12946
Buchbender M, Fehlhofer J, Proff P, Möst T, Ries J, Hannig M, Neurath MF, Gund M, Atreya R, Kesting M (2022) Expression of inflammatory mediators in biofilm samples and clinical association in inflammatory bowel disease patients-a preliminary study. Clin Oral Invest 26:1217–1228. https://doi.org/10.1007/s00784-021-04093-2
Figueredo CM, Brito F, Barros FC, Menegat JS, Pedreira RR, Fischer RG, Gustafsson A (2011) Expression of cytokines in the gingival crevicular fluid and serum from patients with inflammatory bowel disease and untreated chronic periodontitis. J Periodontal Res 46:141–146. https://doi.org/10.1111/j.1600-0765.2010.01303.x
Said H, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, Mano S, Morita H, Dohi T, Oota H, Hattori M (2014) Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 21:15–25. https://doi.org/10.1093/dnares/dst037
Enver A, Ozmeric N, Isler SC, Toruner M, Fidan C, Demirci G, Elgun S, Silva APBD (2022) Evaluation of periodontal status and cytokine levels in saliva and gingival crevicular fluid of patients with inflammatory bowel diseases. J Periodontol 93:1649–1660. https://doi.org/10.1002/JPER.22-0065
Figueredo CM, Martins AP, Lira-Junior R, Menegat JB, Carvalho AT, Fischer RG, Gustafsson A (2017) Activity of inflammatory bowel disease influences the expression of cytokines in gingival tissue. Cytokine 95:1–6. https://doi.org/10.1016/j.cyto.2017.01.016
Gursoy UK, Könönen E (2012) Understanding the roles of gingival beta-defensins. J Oral Microbiol, 4. https://doi.org/10.3402/jom.v4i0.15127
Bentley RW, Pearson J, Gearry RB, Barclay ML, McKinney C, Merriman TR, Roberts RL (2010) Association of higher DEFB4 genomic copy number with Crohn’s disease. Am J Gastroenterol 105:354–359. https://doi.org/10.1038/ajg.2009.582
Aldhous MC, Abu Bakar S, Prescott NJ, Palla R, Soo K, Mansfield JC, Mathew CG, Satsangi J, Armour JA (2010) Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn’s disease. Hum Mol Genet 19:4930–4938. https://doi.org/10.1093/hmg/ddq411
Polley S, Prescott N, Nimmo E, Veal C, Vind I, Munkholm P, Fode P, Mansfield J, Skyt Andersen P, Satsangi JG, Mathew C, Hollox EJ (2016) Copy number variation of scavenger-receptor cysteine-rich domains within DMBT1 and Crohn’s disease. Eur J Hum Genet 24:1294–1300. https://doi.org/10.1038/ejhg.2015.280
Bertl K, Tsakos G, Pandis N, Bogren A, Burisch J, Stavropoulos A (2023) Health-related quality of life aspects of the ‘Periodontitis prevalence in ulcerative colitis and Crohn’s disease’ (PPCC) cohort. J Clin Periodontol Sep 5 [Online ahead of print.] https://doi.org/10.1111/jcpe.1386
Acknowledgements
Katja Sampalahti is acknowledged for her technical assistance during the laboratory analysis.
Funding
Open Access funding provided by University of Turku (including Turku University Central Hospital). This work was funded by the Finnish Cultural Foundation (J.R.) and partly supported by the Institute of Dentistry, University of Turku (V.L.)
Author information
Authors and Affiliations
Contributions
Conceptualization: U.K.G., M.G., and J.R.; methodology: U.K.G., M.G., J.R., and V.L.; investigation: J.R. and M.G.; resources: J.R. and V.L.; writing—original draft preparation: M.G. and U.K.G.; writing—review and editing: V.L. and J.R.; supervision: U.K.G.; project administration: M.G.; funding acquisition: J.R.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Ethical Committee of the Hospital District of Southwest Finland approved the study protocols (114/1801/2016), and the study protocols followed World Medical Association Declaration of Helsinki guidelines. All participants received written information and verbal clarification on study protocols and purposes, and all participants gave their written consent [14].
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gürsoy, U.K., Gürsoy, M., Loimaranta, V. et al. Salivary Th17 cytokine, human β-defensin 1–3, and salivary scavenger and agglutinin levels in Crohn’s disease. Clin Oral Invest 28, 108 (2024). https://doi.org/10.1007/s00784-024-05509-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00784-024-05509-5