Abstract
Objectives
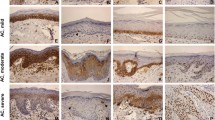
This study aimed to analyze the expression of miR-181b, miR-21, miR-31, and miR-345 in actinic cheilitis with and without epithelial dysplasia and lower lip squamous cell carcinomas, and to verify if the deregulated expression of these miRNAs would be indicative of malignant transformation.
Materials and methods
The sample was selected from formalin-fixed paraffin-embedded tissues of 19 actinic cheilitis without epithelial dysplasia, 32 actinic cheilitis with epithelial dysplasia, 42 lower lip squamous cell carcinomas, and 10 nonaltered oral mucosa of the lip. The microRNA (miR, miRNA) expression was quantified by real-time RT-PCR and the expression of the selected miRNAs among the groups of actinic cheilitis and lower lip cancer was compared by chi-square.
Results
A higher expression of miR-181b, miR-31, and miR-345 was found in actinic cheilitis without epithelial dysplasia in comparison to that in actinic cheilitis with epithelial dysplasia and with lower lip cancer. There were no differences in miR-21 expression between actinic cheilitis and lower lip cancer. Hierarchical clustering analysis showed a tendency for a downregulation of miR-181b, miR-21, miR-31, and miR-345 in most patients with lower lip cancers.
Conclusions
The upregulation of miR-181b, miR-31, and miR-345 expression in actinic cheilitis without epithelial dysplasia and the decrease in the expression of these miRNAs in actinic cheilitis with epithelial dysplasia and in lower lip cancer are potential biomarkers of malignant progression.
Clinical relevance
This miRNA signature can help to identify actinic cheilitis with potential to progress to lip cancer.



Similar content being viewed by others
References
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Courthod G, Franco P, Palermo L, Pisconti S, Numico G (2014) The role of microRNA in head and neck cancer: current knowledge and perspectives. Molecules 19(5):5704–5716. https://doi.org/10.3390/molecules19055704
Chattopadhyay E, De Sarkar N, Singh R, Ray A, Roy R, Paul RR, Pal M, Ghose S, Ghosh S, Kabiraj D, Banerjee R, Roy B (2016) Genome-wide mitochondrial DNA sequence variations and lower expression of OXPHOS genes predict mitochondrial dysfunction in oral cancer tissue. Tumour Biol 37(9):11861–11871. https://doi.org/10.1007/s13277-016-5026-x
Cao M, Zheng L, Liu J, Dobleman T, Hu S, Go VLW, Gao G, Xiao GG (2018) MicroRNAs as effective surrogate biomarkers for early diagnosis of oral cancer. Clin Oral Investig 22(2):571–581. https://doi.org/10.1007/s00784-017-2317-6
Min A, Zhu C, Peng S, Rajthala S, Costea DE, Sapkota D (2015) MicroRNAs as important players and biomarkers in oral carcinogenesis. Biomed Res Int 2015:186904. https://doi.org/10.1155/2015/186904
Ramassone A, Pagotto S, Veronese A, Visone R (2018) Epigenetics and MicroRNAs in cancer. Int J Mol Sci 19(2). https://doi.org/10.3390/ijms19020459
Maclellan SA, Lawson J, Baik J, Guillaud M, Poh CF, Garnis C (2012) Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med 1(2):268–274. https://doi.org/10.1002/cam4.17
Zhang H, Li T, Zheng L, Huang X (2017) Biomarker MicroRNAs for diagnosis of oral squamous cell carcinoma identified based on gene expression data and MicroRNA-mRNA network analysis. Comput Math Methods Med 2017:9803018. https://doi.org/10.1155/2017/9803018
Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo Muzio L (2018) Predictive prognostic value of tissue-based microRNA expression in oral squamous cell carcinoma: a systematic review and meta-analysis. J Dent Res 97(7):759–766. https://doi.org/10.1177/0022034518762090
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC (2010) Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis 16(4):360–364. https://doi.org/10.1111/j.1601-0825.2009.01646.x
El-Sakka H, Kujan O, Farah CS (2018) Assessing miRNAs profile expression as a risk stratification biomarker in oral potentially malignant disorders: a systematic review. Oral Oncol 77:57–82. https://doi.org/10.1016/j.oraloncology.2017.11.021
Gale NP, Sidransky D, Westra W, Califano J (2005) Epithelial precursor lesions. In: Barnes L, Eveson JW, Reichart P, Sidransky D, et al. (eds). In: Press I (ed) World Health Organization Classification of Tumours. Pathology and Genetics. Head and Neck Tumours. Lyon, France, pp 140–143
Reibel J, Gale N, Hille J, Hunt JL, Lingen M, Muller S et al (2017) Oral potentially malignant disorders and oral epithelial dysplasia. In: ElNaggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ (eds) WHO Classification of head and neck tumours, 4th edn. International Agency for Research on Cancer, Lyon, pp 112–115
Speight PM, Abram TJ, Floriano PN et al (2015) Interobserver agreement in dysplasia grading: toward an enhanced gold standard for clinical pathology trials. Oral Surg Oral Med Oral Pathol Oral Radiol 120(4):474–482. https://doi.org/10.1016/j.oooo.2015.05.023
Nagata G, Santana T, Queiroz A, Caramez RH, Trierveiler M (2018) Evaluation of epithelial dysplasia adjacent to lip squamous cell carcinoma indicates that the degree of dysplasia is not associated with the occurrence of invasive carcinoma in this site. J Cutan Pathol. https://doi.org/10.1111/cup.13270
Pilati S, Bianco BC, Vieira D, Modolo F (2017) Histopathologic features in actinic cheilitis by the comparison of grading dysplasia systems. Oral Dis 23(2):219–224. https://doi.org/10.1111/odi.12597
Cervigne NK, Reis PP, Machado J, Sadikovic B, Bradley G, Galloni NN, Pintilie M, Jurisica I, Perez-Ordonez B, Gilbert R, Gullane P, Irish J, Kamel-Reid S (2009) Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet 18(24):4818–4829. https://doi.org/10.1093/hmg/ddp446
Xiao W, Bao ZX, Zhang CY, Zhang XY, Shi LJ, Zhou ZT, Jiang WW (2012) Upregulation of miR-31* is negatively associated with recurrent/newly formed oral leukoplakia. PLoS One 7(6):e38648. https://doi.org/10.1371/journal.pone.0038648
Yang Y, Li YX, Yang X, Jiang L, Zhou ZJ, Zhu YQ (2013) Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. BMC Cancer 13:129. https://doi.org/10.1186/1471-2407-13-129
Brito JA, Gomes CC, Guimaraes AL, Campos K, Gomez RS (2014) Relationship between microRNA expression levels and histopathological features of dysplasia in oral leukoplakia. J Oral Pathol Med 43(3):211–216. https://doi.org/10.1111/jop.12112
Maimaiti A, Abudoukeremu K, Tie L, Pan Y, Li X (2015) MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene 558(2):271–277. https://doi.org/10.1016/j.gene.2015.01.004
Zhu G, He Y, Yang S, Chen B, Zhou M, Xu XJ (2015, 2015) Identification of gene and MicroRNA signatures for oral cancer developed from oral leukoplakia. Biomed Res Int:841956. https://doi.org/10.1155/2015/841956
Hung KF, Liu CJ, Chiu PC, Lin JS, Chang KW, Shih WY, Kao SY, Tu HF (2016) MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol 53:42–47. https://doi.org/10.1016/j.oraloncology.2015.11.017
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Brito BL, Lourenco SV, Damascena AS, Kowalski LP, Soares FA, Coutinho-Camillo CM (2016) Expression of stem cell-regulating miRNAs in oral cavity and oropharynx squamous cell carcinoma. J Oral Pathol Med 45(9):647–654. https://doi.org/10.1111/jop.12424
Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH, Cheng HW, Lin SC (2011) miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med 40(5):397–404. https://doi.org/10.1111/j.1600-0714.2010.01003.x
Deng ZY, Wang YH, Quan HZ, Liu OS, Li YP, Li Y, Zhu W, Munnee K, Tang ZG (2016) Investigation of the association between miR181b, Bcl2 and LRIG1 in oral verrucous carcinoma. Mol Med Rep 14(4):2991–2996. https://doi.org/10.3892/mmr.2016.5608
Yin J, Shi Z, Wei W, Lu C, Wei Y, Yan W, Li R, Zhang J, You Y, Wang X (2020) MiR-181b suppress glioblastoma multiforme growth through inhibition of SP1-mediated glucose metabolism. Cancer Cell Int 20:69. https://doi.org/10.1186/s12935-020-1149-7
Lin Z, Li D, Cheng W, Wu J, Wang K, Hu Y (2019) MicroRNA-181 Functions as an antioncogene and mediates NF-kappaB pathway by targeting RTKN2 in ovarian cancers. Reprod Sci 26(8):1071–1081. https://doi.org/10.1177/1933719118805865
Funding
This study was supported Brazilian National Council for Scientific and Technological Development (CNPq) grant: #870891/1999-5 and partially supported by “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil- CAPES,” financial code #001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The Committee of Ethics and Research of the Bauru School of Dentistry, University of São Paulo, has approved this study (process #: 1.929.331 – CAAE 63879016.5.0000.5417).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Assao, A., Domingues, M.A.C., Minicucci, E.M. et al. The relevance of miRNAs as promising biomarkers in lip cancer. Clin Oral Invest 25, 4591–4598 (2021). https://doi.org/10.1007/s00784-020-03773-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-020-03773-9