Abstract
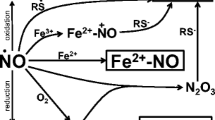
Protein arginine N-methyltransferases (PRMTs) have emerged as important actors in the eukaryotic stress response with implications in human disease, aging, and cell signaling. Intracellular free methylarginines contribute to cellular stress through their interaction with nitric oxide synthase (NOS). The arginine-dependent production of nitric oxide (NO), which is strongly inhibited by methylarginines, serves as a protective small molecule against oxidative stress in eukaryotic cells. NO signaling is highly conserved between higher and lower eukaryotes, although a canonical NOS homologue has yet to be identified in yeast. Since stress signaling pathways are well conserved among eukaryotes, yeast is an ideal model organism to study the implications of PRMTs and methylarginines during stress. We sought to explore the roles and fates of methylarginines in Saccharomyces cerevisiae. We starved methyltransferase-, autophagy-, and permease-related yeast knockouts by incubating them in water and monitored methylarginine production. We found that under starvation, methylarginines are expelled from yeast cells. We found that autophagy-deficient cells have an impaired ability to efflux methylarginines, which suggests that methylarginine-containing proteins are degraded via autophagy. For the first time, we determine that yeast take up methylarginines less readily than arginine, and we show that methylarginines impact yeast NO production. This study reveals that yeast circumvent a potential methylarginine toxicity by expelling them after autophagic degradation of arginine-modified proteins.










Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file.
Abbreviations
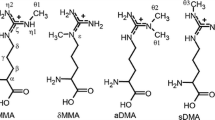
- aDMA:
-
Nη,Nη-asymmetric dimethylarginine
- Atg:
-
Autophagy-related protein
- DAF-FM DA:
-
4-amino-5-methylamino-2',7'-difluorofluorescein diacetate
- DDAH:
-
Dimethylarginine dimethylaminohydrolase
- GST-GAR:
-
GST-tagged glycine and arginine-rich polypeptide
- KO:
-
Knockout
- MMA:
-
Monomethylarginine
- NO:
-
Nitric oxide
- NOS:
-
Nitric oxide synthase
- OD:
-
Optical density
- PRMT:
-
Protein arginine N-methyltransferase
- SC:
-
Synthetic complete media
- sDMA:
-
Nη1,Nη2-symmetric dimethylarginine
- YPD:
-
Yeast–peptone–dextrose media
References
Ahmad M, Bussey H (1986) Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr Genet 10:587–592. https://doi.org/10.1007/BF00418125
Almeida B, Buttner S, Ohlmeier S et al (2007) NO-mediated apoptosis in yeast. J Cell Sci 120:3279–3288. https://doi.org/10.1242/jcs.010926
Astuti RI, Nasuno R, Takagi H (2016) Nitric oxide signaling in yeast. Appl Microbiol Biotechnol 100:9483–9497. https://doi.org/10.1007/s00253-016-7827-7
Astuti RI, Watanabe D, Takagi H (2016) Nitric oxide signaling and its role in oxidative stress response in Schizosaccharomyces pombe. Nitric Oxide - Biol Chem 52:29–40. https://doi.org/10.1016/j.niox.2015.11.001
Babu BR, Frey C, Griffith OW (1999) L-arginine binding to nitric-oxide synthase. The role of H-bonds to the nonreactive guanidinium nitrogens. J Biol Chem 274:25218–25226. https://doi.org/10.1074/jbc.274.36.25218
Bassareo PP, Mussap M, Bassareo V et al (2015) Nitrergic system and plasmatic methylarginines: Evidence of their role in the perinatal programming of cardiovascular diseases. Clin Chim Acta 451:21–27. https://doi.org/10.1016/j.cca.2015.05.011
Blanc RS, Richard S (2017) Arginine methylation: the coming of age. Mol Cell 65:8–24. https://doi.org/10.1016/j.molcel.2016.11.003
Bogle RG, MacAllister RJ, Whitley GSJ, Vallance P (1995) Induction of N(G)-monomethyl-L-arginine uptake: A mechanism for differential inhibition of NO synthases? Am J Physiol - Cell Physiol 269:C750–C756. https://doi.org/10.1152/ajpcell.1995.269.3.c750
Boisvert F-M, Déry U, Masson J-Y, Richard S (2005) Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev 19:671–676. https://doi.org/10.1101/gad.1279805
Brüne B, Von Knethen A, Sandau KB (1998) Nitric oxide and its role in apoptosis. Eur J Pharmacol 351:261–272. https://doi.org/10.1016/S0014-2999(98)00274-X
Cardounel AJ, Cui H, Samouilov A et al (2007) Evidence for the pathophysiological role of endogenous methylarginines in regulation of endothelial no production and vascular function. J Biol Chem 282:879–887. https://doi.org/10.1074/jbc.M603606200
Carilla-Latorre S, Annesley SJ, Munoz-Braceras S et al (2013) Ndufaf5 deficiency in the Dictyostelium model: new roles in autophagy and development. Mol Biol Cell 24:1519–1528. https://doi.org/10.1091/mbc.E12-11-0796
Cebollero E, Reggiori F (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta - Mol Cell Res 1793:1413–1421. https://doi.org/10.1016/j.bbamcr.2009.01.008
Conway EM, Carmeliet P (2004) The diversity of endothelial cells: a challenge for therapeutic angiogenesis. Genome Biol 5:207. https://doi.org/10.1186/gb-2004-5-2-207
Gary JD, Lin WJ, Yang MC et al (1996) The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem 271:12585–12594. https://doi.org/10.1074/jbc.271.21.12585
Giaever G, Nislow C (2014) The yeast deletion collection: a decade of functional genomics. Genetics 197:451–465. https://doi.org/10.1534/genetics.114.161620
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34. https://doi.org/10.1007/978-1-4939-1363-3_1
Gu H, Liu G, Wang J et al (2014) Selecting the correct weighting factors for linear and quadratic calibration curves with least-squares regression algorithm in bioanalytical LC-MS/MS assays and impacts of using incorrect weighting factors on curve stability, data quality, and assay perfo. Anal Chem 86:8959–8966. https://doi.org/10.1021/ac5018265
Guan J, Stromhaug PE, George MD et al (2001) Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell 12:3821–3838
Haghandish N, Baldwin RM, Morettin A et al (2019) PRMT7 methylates eukaryotic translation initiation factor 2α and regulates its role in stress granule formation. Mol Biol Cell 30:778–793. https://doi.org/10.1091/mbc.E18-05-0330
Hardwick JS, Kuruvilla FG, Tong JK et al (1999) Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci 96:14866–14870
Huang WP, Scott SV, Kim J, Klionsky DJ (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275:5845–5851. https://doi.org/10.1074/jbc.275.8.5845
Huang L, Wang Z, Narayanan N, Yang Y (2018) Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res 46:3061–3074. https://doi.org/10.1093/nar/gky103
Isnard AD, Thomas D, Surdin-Kerjan Y (1996) The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J Mol Biol 262:473–484. https://doi.org/10.1006/jmbi.1996.0529
Kawano-kawada M, Kakinuma Y, Sekito T (2018) Transport of amino acids across the vacuolar membrane of yeast : Its mechanism and physiological role. Biol Pharm Bull 41:1496–1501
Kerstens W, Van Dijck P, Leuven K (2018) A Cinderella story: how the vacuolar proteases Pep4 and Prb1 do more than cleaning up the cell’s mass degradation processes. Microb Cell. 5:438–443. https://doi.org/10.15698/mic2018.10.650
Kimoto M, Miyatake S, Sasagawa T et al (1998) Purification, cDNA cloning and expression of human N(G), N(G)- dimethylarginine dimethylaminohydrolase. Eur J Biochem 258:863–868. https://doi.org/10.1046/j.1432-1327.1998.2580863.x
Kojima H, Sakurai K, Kikuchi K et al (1998) Development of a fluorescent indicator for nitric oxide based on the fluorescein chromophore. Chem Pharm Bull 46:373–375
Kunz HE, Dorschner JM, Berent TE et al (2020) Methylarginine metabolites are associated with attenuated muscle protein synthesis in cancer-associated muscle wasting. J Biol Chem 295:17441–17459. https://doi.org/10.1074/jbc.RA120.014884
Lakowski TM, Szeitz A, Pak ML et al (2013) MS3 fragmentation patterns of monomethylarginine species and the quantification of all methylarginine species in yeast using MRM3. J Proteomics 80:43–54. https://doi.org/10.1016/j.jprot.2013.01.003
Lakowski TM, Pak ML, Szeitz A et al (2015) Arginine methylation in yeast proteins during stationary-phase growth and heat shock. Amino Acids 47:2561–2571. https://doi.org/10.1007/s00726-015-2047-5
Larimer FW, Ramey W, Lijinsky W, Epler JL (1978) Mutagenicity of methylated N-nitrosopiperidines in Saccharomyces cerevisiae. Mutat Res 57:155–161
Leiper J, Vallance P (1999) Biological significance of endogenous methylarginines that inhibit nitric oxide synthases. Cardiovasc Res 43:542–548. https://doi.org/10.1016/S0008-6363(99)00162-5
Li B, Skinner C, Castello PR et al (2011) Identification of potential calorie restriction-mimicking yeast mutants with increased mitochondrial respiratory chain and nitric oxide levels. J Aging Res 2011:673185. https://doi.org/10.4061/2011/673185
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 130:165–178. https://doi.org/10.1016/j.cell.2007.05.021
Nasuno R, Aitoku M, Manago Y et al (2014) Nitric oxide-mediated antioxidative mechanism in yeast through the activation of the transcription factor Mac1. PLoS One 9:e113788. https://doi.org/10.1371/journal.pone.0113788
Nasuno R, Yoshikawa Y, Takagi H (2022) Acetaldehyde reacts with a fluorescent nitric oxide probe harboring an o-phenylenediamine structure that interferes with fluorometry. Free Radic Biol Med 187:29–37. https://doi.org/10.1016/j.freeradbiomed.2022.05.014
Niewmierzycka A, Clarke S (1999) S -Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J Biol Chem 274:814–824
Nishimura A, Kawahara N, Takagi H (2013) The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem Biophys Res Commun 430:137–143. https://doi.org/10.1016/j.bbrc.2012.11.023
Olken NM, Marletta MA (1993) NG-Methyl-l-arginine functions as an alternate substrate and mechanism-based inhibitor of nitric oxide synthase. Biochemistry 32:9677–9685. https://doi.org/10.1021/bi00088a020
Opekarová M, Kubín J (1997) On the unidirectionality of arginine uptake in the yeast Saccharomyces cerevisiae. FEMS Microbiol Lett 152:261–267. https://doi.org/10.1016/S0378-1097(97)00207-3
Osorio NS, Carvalho A, Almeida AJ et al (2007) Nitric oxide signaling is disrupted in the yeast model for batten disease. Mol Biol Cell 18:2755–2767. https://doi.org/10.1091/mbc.E06
Pahlich S, Zakaryan RP, Gehring H (2006) Protein arginine methylation: cellular functions and methods of analysis. Biochim Biophys Acta Proteins Proteom 1764:1890–1903. https://doi.org/10.1016/j.bbapap.2006.08.008
Prinsen HCMT, Schiebergen-Bronkhorst BGM, Roeleveld MW et al (2016) Rapid quantification of underivatized amino acids in plasma by hydrophilic interaction liquid chromatography (HILIC) coupled with tandem mass-spectrometry. J Inherit Metab Dis 39:651–660. https://doi.org/10.1007/s10545-016-9935-z
Regenberg B, Düring-Olsen L, Kielland-Brandt MC, Holmberg S (1999) Substrate specificity and gene expression of the amino acid permeases in Saccharomyces cerevisiae. Curr Genet 36:317–328. https://doi.org/10.1007/s002940050506
Sayegh J, Clarke SG (2008) Hsl7 is a substrate-specific type II protein arginine methyltransferase in yeast. Biochem Biophys Res Commun 372:811–815. https://doi.org/10.1016/j.bbrc.2008.05.121
Sherman F (2002) Getting Started with Yeast. Methods Enzymol 350:3–41. https://doi.org/10.1016/S0076-6879(02)50954-X
Shirakawa T, Kako K, Shimada T et al (2011) Production of free methylarginines via the proteasome and autophagy pathways in cultured cells. Mol Med Rep 4:615–620. https://doi.org/10.3892/mmr.2011.488
Staszek P, Krasuska U, Otulak-Kozieł K et al (2019) Canavanine-induced decrease in nitric oxide synthesis alters activity of antioxidant system but does not impact S-nitrosoglutathione catabolism in tomato roots. Front Plant Sci 10:1077. https://doi.org/10.3389/fpls.2019.01077
Teste MA, Duquenne M, François JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99. https://doi.org/10.1186/1471-2199-10-99
Tojo A, Welch WJ, Bremer V et al (1997) Colocalization of demethylating enzymes and NOS and functional effects of methylarginines in rat kidney. Kidney Int 52:1593–1601. https://doi.org/10.1038/ki.1997.490
Torggler R, Papinski D, Kraft C (2017) Assays to monitor autophagy in Saccharomyces cerevisiae. Cells 6:23. https://doi.org/10.3390/cells6030023
Urano J, Tabancay AP, Yang W, Tamanoi F (2000) The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J Biol Chem 275:11198–11206. https://doi.org/10.1074/jbc.275.15.11198
Vallance P, Leone A, Calver A, Collier JMS (1992) Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 339:572–575
Wang H, Wang H, Huang ZQ et al (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853–857. https://doi.org/10.1126/science.1060781
Xie W, Denman RB (2011) Protein methylation and stress granules: posttranslational remodeler or innocent bystander? Mol Biol Int 2011:137459. https://doi.org/10.4061/2011/137459
Yang Z, Klionsky DJ (2007) Permeases recycle amino acids resulting from autophagy. Autophagy 3:149–150. https://doi.org/10.4161/auto.3631
Yang Z, Huang J, Geng J et al (2006) Atg22 recycles amino acids to link the degradative and recycling functions of autophagy zhifen. Mol Biol Cell 17:5094–5104. https://doi.org/10.1091/mbc.E06
Young BD, Weiss DI, Zurita-Lopez CI et al (2012) Identification of methylated proteins in the yeast small ribosomal subunit: A role for SPOUT methyltransferases in protein arginine methylation. Biochemistry 51:5091–5104. https://doi.org/10.1021/bi300186g
Acknowledgements
The authors thank Dr. Corey Nislow and technical support from Jennifer Chiang for the generous donation of several yeast strains used in this study. The authors thank Dr. Karla Williams for use of her microscope. The authors thank Dr. Colin Ross and his team for their support in planning RNA quantification experiments. We also thank Fraser Ball and Diana Vasileva for their early contributions to this project.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) RGPIN-2015-04450 and RGPIN-2020-04227 Discovery Grants (A.F.), NSERC CGS-M (J.I.B), NSERC CGS-D (J.I.B.), and the NSERC Undergraduate Student Research Award (E.Z.).
Author information
Authors and Affiliations
Contributions
AF and JIB conceptualized the study and methodologies. JIB performed the research, analyzed the data, and wrote the paper. JA and EZ performed experiments with guidance from JIB and contributed methods to the paper. AF and JIB edited the final draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests with the contents of this article.
Additional information
Handling editor: J. Metcalf.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Brown, J.I., Alibhai, J., Zhu, E. et al. Methylarginine efflux in nutrient-deprived yeast mitigates disruption of nitric oxide synthesis. Amino Acids 55, 215–233 (2023). https://doi.org/10.1007/s00726-022-03220-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-022-03220-x