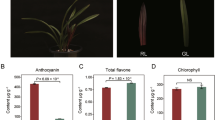
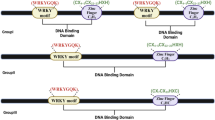
Abstract
Downy mildew, resulted from Plasmopara viticola, is one of most severe fungal diseases of grapevine. Since Vitis vinifera is susceptible to downy mildew, much effort has been focused on improving the resistance of V. vinifera. The Chinese wild V. pseudoreticulata accession Baihe-35-1 (BH) shows resistance to P. viticola; however, the molecular mechanism underlying its resistance to P. viticola is largely unknown. In order to better understand the cellular processes, the transcriptomic changes were investigated at 0, 12, 24, 48, 96, and 120 h post infection (hpi). Transcriptome analysis identified a total of 175 differentially expressed genes. Most of them were found to be associated with oxidative stress, cell wall modification, and protein modification. Moreover, the BH resistance to P. viticola was involved in metabolism process, including terpene synthesis and hormone synthesis. In addition, we verified 12 genes to ensure the accuracy of transcriptome data using quantitative real-time PCR (qRT-PCR). This study broadly characterizes a molecular mechanism in which oxidative stress and cell wall biosynthesis and modification play important roles in the response of BH to P. viticola and provides a basis for further analysis of key genes involved in the resistance to P. viticola.







Similar content being viewed by others
Data availability
The data supporting the conclusions of this article are within the paper and its additional files.
Abbreviations
- BH:
-
V. pseudoreticulata accession Baihe-35-1
- PR:
-
Pathogenesis-related protein
- ROS:
-
Reactive oxygen species
- HR:
-
Hypersensitive response
- NPR1:
-
Nonexpresser of PR genes 1
- TLP:
-
Thaumatin-like protein
- PUB:
-
Plant U-box
- VDAC:
-
Voltage-dependent anion channel
- CPM:
-
Counts per millions
- DEG:
-
Differentially expressed gene
- GO:
-
Gene Ontology
- RPKM:
-
Reads Per Kilobase per Million of mapped reads
- ceQORH:
-
Chloroplast envelope quinone oxidoreductase homolog
- GSTU17:
-
Glutathione S-transferase U17
- CRL1:
-
Cinnamoyl CoA:NADP oxidoreductase-like 1
- GIM2:
-
Germination insensitive to ABA mutant 2
- PCO4:
-
Plant cysteine oxidase 4
- PME17:
-
Pectin methylesterase inhibitor 17
- BXL1:
-
Beta-d-xylosidase 1
- F5H:
-
Ferulate 5-hydroxylase
- PRX52:
-
Peroxidase 52
- CESA6:
-
Cellulose synthase 6
- XTH33:
-
Xyloglucan endotransglucosylase/hydrolase 33
- SAUR39:
-
Small auxin upregulated RNA 39
- PIN7:
-
PIN-formed 7
- CYP707A1:
-
Cytochrome p450, family 707, subfamily A, polypeptide 1
- ACO1:
-
1-Aminocyclopropane-1-carboxylate oxidase 1
- TF:
-
Transcription factor
- ASMT:
-
N-acetylserotonin O-methyltransferase
- UGT:
-
UDP-glucosyl transferase
- FKBP:
-
FK506 binding protein
- RHA2A:
-
RING-H2 A2A
- NRT1:
-
Nitrate transporter 1
- AZF1:
-
Zinc-finger protein 1
References
Alarcon AA, Lazazzara V, Cappellin L, Bianchedi PL, Schuhmacher R, Wohlfahrt G, Pertot I, Biasioli F, Perazzolli M (2015) Emission of volatile sesquiterpenes and monoterpenes in grapevine genotypes following Plasmopara viticola inoculation in vitro. J Mass Spectrom 50:1013–1022
Alberto AP, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54:263–272
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120:163–176
Burruano S (2000) The life-cycle of Plasmopara viticola, cause of downy mildew of vine. Mycologist 14:179–182
Carisse O (2016) Development of grape downy mildew (Plasmopara viticola) under northern viticulture conditions: influence of fall disease incidence. Eur J Plant Pathol 144:773–783
Cheng AX, Lou YG, Mao YB, Lu S, Wang LJ, Chen XY (2007) Plant terpenoids: biosynthesis and ecological functions. J Integr Plant Biol 49:179–186
Curien G, Giustini C, Montillet JL, Mas-y-Mas S, Cobessi D, Ferrer JL, Matringe M, Grechkin A, Rolland N (2016) The chloroplast membrane associated ceQORH putative quinone oxidoreductase reduces long-chain, stress-related oxidized lipids. Phytochemistry 122:45–55
de Vega D, Newton AC, Sadanandom A (2018) Post-translational modifications in priming the plant immune system: ripe for exploitation? FEBS Lett 592:1929–1936
Diez-Navajas A, Wiedemann-Merdinoglu S, Greif C, Merdinoglu D (2008) Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 98:776–780
Divilov K, Barba P, Cadle-Davidson L, Reisch BI (2018) Single and multiple phenotype QTL analyses of downy mildew resistance in interspecific grapevines. Theor Appl Genet 131:1133–1143
Dmitriev AA, Krasnov GS, Rozhmina TA, Kishlyan NV, Zyablitsin AV, Sadritdinova AF, Snezhkina AV, Fedorova MS, Yurkevich OY, Muravenko OV, Bolsheva NL, Kudryavtseva AV, Melnikova NV (2016) Glutathione S-transferases and UDP-glycosyltransferases are involved in response to aluminum stress in flax. Front Plant Sci 7:1920
Fan G, Yang Y, Li T, Lu W, Du Y, Qiang X, Wen Q, Shan W (2018) A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum-mediated immunity. Mol Plant 11:1067–1083
Figueiredo AF, Margarida A, Ferreira S, Sebastiana M, Choi YH, Sousa L, Acioli-Santos B, Pessoa F, Verpoorte R, Pais MS (2008) Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J Exp Bot 59:3371–3381
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Töpfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108:501–515
Fröbel S, Dudenhöffer J, Töpfer R, Zyprian E (2019) Transcriptome analysis of early downy mildew (Plasmopara viticola) defense in grapevines carrying the Asian resistance locus Rpv10. Euphytica 215:28
Gabaston J, Richard T, Biais B, Waffo-Teguo P, Pedrot E, Jourdes M, Corio-Costet MF, Mérillon JM (2017) Stilbenes from common spruce (Picea abies) bark as natural antifungal agent against downy mildew (Plasmopara viticola). Ind Crop Prod 103:267–273
Gessler C, Pertot I, Perazzolli M (2011) Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol Mediterr 50:3–44
Gindro K, Pezet R, Viret O (2003) Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol Biochem 41:846–853
Gong P, Riemann M, Dong D, Stoeffler N, Gross B, Markel A, Nick P (2019) Two grapevine metacaspase genes mediate ETI-like cell death in grapevine defence against infection of Plasmopara viticola. Protoplasma:1–19. https://doi.org/10.1007/s00709-019-01353-7
He R, Wu J, Zhang Y, Agüero CB, Li X, Liu S, Wang C, Walker MA, Lu J (2016) Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma 254:1579–1589
Henanff GL, Heitz T, Mestre P, Mutterer J, Walter B, Chong J (2009) Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol 9:54
Huang X, Zhu G, Liu Q, Chen L, Li Y, Hou B (2018) Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoic acids. Plant Physiol 176:3103–3119
Jürges G, Kassemeyer HH, Dürrenberger M, Düggelin M, Nick P (2009) The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol 11:886–898
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:1
Kortekamp A, Zyprian E (1999) Leaf hairs as a basic protective barrier against downy mildew of grape. J Phytopathol 147:453–459
Kortekamp A, Zyprian E (2003) Characterization of Plasmopara-resistance in grapevine using in vitro plants. J Plant Physiol 160:1393–1400
Lazazzara V, Bueschl C, Parich A, Pertot I, Schuhmacher R, Perazzolli M (2018) Downy mildew symptoms on grapevines can be reduced by volatile organic compounds of resistant genotypes. Sci Rep 8:1618
Lenzi L, Caruso C, Bianchedi PL, Pertot I, Perazzolli M (2016) Laser microdissection of grapevine leaves reveals site-specific regulation of transcriptional response to Plasmopara viticola. Plant Cell Physiol 57:69–81
Li X, Wu J, Yin L, Zhang Y, Qu J, Lu J (2015) Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol Biochem 95:1–14
Li J, Zhang Y, Jiang L (2016) Overexpression of a stress-responsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘Thompson Seedless’. Plant Physiol Biochem 112:53–63
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu R, Wang L, Zhu J, Chen T, Wang Y, Xu Y (2015) Histological responses to downy mildew in resistant and susceptible grapevines. Protoplasma 252:259–270
Ma H, Xiang G, Li Z, Wang Y, Dou M, Su L, Yin X, Liu R, Wang Y, Xu Y (2018) Grapevine VpPR10.1 functions in resistance to Plasmopara viticola through triggering a cell death-like defense response by interacting with VpVDAC3. Plant Biotechnol J 16:1488–1501
Malacarne G, Vrhovsek U, Zulini L, Cestaro A, Stefanini M, Mattivi F, Delledonne M, Velasco R, Moser C (2011) Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol 11:114
Marchive C, Léon C, Kappel C, Coutos-Thévenot P, Corio-Costet MF, Delrot S, Lauvergeat V (2013) Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS One 8:e54185
Merdinoglu D, Wiedeman-Merdinoglu S, Coste P, Dumas V, Haetty S, Butterlin G, Greif C (2003) Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hortic 603:451–456
Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5:358
Milli A, Cecconi D, Bortesi L, Persi A, Rinalducci S, Zamboni A, Zoccatelli G, Lovato A, Zolla L, Polverari A (2012) Proteomic analysis of the compatible interaction between Vitis vinifera and Plasmopara viticola. J Proteome 75:1284–1302
Moreira FM, Madini A, Marino R, Zulini L, Stefanini M, Velasco R, Kozma P, Grando MS (2011) Genetic linkage maps of two interspecific grape crosses (Vitis spp.) used to localize quantitative trait loci for downy mildew resistance. Tree Genet Genomes 7:153–167
Nascimento R, Maia M, Ferreira AEN, Silva AB, Freire AP, Cordeiro C, Silva MS, Figueiredo A (2019) Early stage metabolic events associated with the establishment of Vitis vinifera - Plasmopara viticola compatible interaction. Plant Physiol Biochem 137:1–13
Nascimento-Gavioli MCA, Agapito-Tenfen SZ, Nodari RO, Welter LJ, Sanchez Mora FD, Saifert L, da Silva AL, Guerra MP (2017) Proteome of Plasmopara viticola-infected Vitis vinifera provides insights into grapevine Rpv1/Rpv3 pyramided resistance to downy mildew. J Proteome 151:264–274
Ochßner I, Hausmann L, Töpfer R (2016) Rpv14, a new genetic source for Plasmopara viticola resistance conferred by Vitis cinerea. Vitis 55:79–81
Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9:534–540
Pezet R, Perret C, Jean-Denis JB, Tabacchi R, Gindro K, Viret O (2003) δ-Viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. J Agric Food Chem 51:5488–5492
Pezet R, Gindro K, Viret O, Spring JL (2004) Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol Mol Plant Pathol 65:297–303
Polesani M, Bortesi L, Ferrarini A, Zamboni A, Fasoli M, Zadra C, Lovato A, Pezzotti M, Delledonne M, Polverari A (2010) General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics 11:117
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rumbou A, Gessler C (2004) Genetic dissection of Plasmopara viticola population from a Greek vineyard in two consecutive years. Eur J Plant Pathol 110:379–392
Sapkota S, Chen LL, Yang S, Hyma KE, Cadle-Davidson L, Hwang CF (2019) Construction of a high-density linkage map and QTL detection of downy mildew resistance in Vitis aestivalis-derived ‘Norton’. Theor Appl Genet 132:137–147
Schwander F, Eibach R, Fechter I, Hausmann L, Zyprian E, Töpfer R (2012) Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet 124:163–176
Singh B, Sharma RA (2015) Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 5:129–151
Su H, Jiao YT, Wang FF, Liu YE, Niu WL, Liu GT, Xu Y (2018) Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant Cell Rep 37:819–832
Tholl D, Lee S (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9:e0143
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc 7:562–578
Veikondis R, Burger P, Vermeulen A, Van Heerden CJ, Prins R (2018) Confirmation of the effectiveness and genetic positions of disease resistance loci in ‘Kishmish Vatkana’ (Ren1) and ‘Villard Blanc’ (Ren3 and Rpv3). S Afr J Enol Vitic 39:185–195
Venuti S, Copetti D, Foria S, Falginella L, Hoffmann S, Bellin D, Cindrić P, Kozma P, Scalabrin S, Morgante M (2013) Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS One 8:e61228
Vezzulli S, Malacarne G, Masuero D, Vecchione A, Dolzani C, Goremykin V, Mehari ZH, Banchi E, Velasco R, Stefanini M, Vrhovsek U, Zulini L, Franceschi P, Moser C (2019) The Rpv3-3 haplotype and stilbenoid induction mediate downy mildew resistance in a grapevine interspecific population. Front Plant Sci 10:234
Wan Y, Schwaninger H, He P, Wang Y (2007) Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 46:132–136
Wang J, Yao W, Wang L, Ma F, Tong W, Wang C, Bao R, Jiang C, Yang Y, Zhang J, Xu Y, Wang X, Zhang C, Wang Y (2017a) Overexpression of VpEIFP1, a novel F-box/Kelch-repeat protein from wild Chinese Vitis pseudoreticulata, confers higher tolerance to powdery mildew by inducing thioredoxin z proteolysis. Plant Sci 263:142–155
Wang L, Xie X, Yao W, Wang J, Ma F, Wang C, Yang Y, Tong W, Zhang J, Xu Y, Wang X, Zhang C, Wang Y (2017b) RING-H2-type E3 gene VpRH2 from Vitis pseudoreticulata improves resistance to powdery mildew by interacting with VpGRP2A. J Exp Bot 68:1669–1687
Wang C, Wu J, Zhang Y, Lu J (2018a) Muscadinia rotundifolia ‘Noble’ defense response to Plasmopara viticola inoculation by inducing phytohormone-mediated stilbene accumulation. Protoplasma 255:95–107
Wang H, Ma D, Yang J, Deng K, Li M, Ji X, Zhong L, Zhao H (2018b) An integrative volatile terpenoid profiling and transcriptomics analysis for gene mining and functional characterization of AvBPPS and AvPS involved in the monoterpenoid biosynthesis in Amomum villosum. Front Plant Sci 9:846
Welter LJ, Göktürk-Baydar N, Akkurt M, Maul E, Eibach R, Töpfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breed 20:359–374
Wu J, Zhang Y, Zhang H, Huang H, Folta KM, Lu J (2010) Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol 10:234
Xing L, Gao L, Chen Q, Pei H, Di Z, Xiao J, Wang H, Ma L, Chen P, Cao A, Wang X (2018) Over-expressing a UDP-glucosyltransferase gene (Ta-UGT 3 ) enhances Fusarium head blight resistance of wheat. Plant Growth Regul 84:561–571
Xu P, Liu Y, Qin H, Ai J, Fan S, Yang Y, Zhao Y, Li X, Li X (2015) Proteomic analysis of the resistant responses of two Vitis amurensis cultivars to Plasmopara viticola infections. Curr Proteomics 12:63–68
Yin X, Liu RQ, Su H, Su L, Guo YR, Wang ZJ, Du W, Li MJ, Zhang X, Wang YJ, Liu GT, Xu Y (2017) Pathogen development and host responses to Plasmopara viticola in resistant and susceptible grapevines: an ultrastructural study. Hortic Res 4:17033
Yu Y, Xu W, Wang S, Xu Y, Li H, Wang Y, Li S (2011) VpRFP1, a novel C4C4-type RING finger protein gene from Chinese wild Vitis pseudoreticulata, functions as a transcriptional activator in defence response of grapevine. J Exp Bot 62:5671–5682
Yu Y, Zhang Y, Yin L, Lu J (2012) The mode of host resistance to Plasmopara viticola infection of grapevines. Phytopathology 102:1094–1101
Zyprian E, Ochssner I, Schwander F, Simon S, Hausmann L, Bonow-Rex M, Moreno-Sanz P, Grando MS, Wiedemann-Merdinoglu S, Merdinoglu D, Eibach R, Toepfer R (2016) Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol Gen Genomics 291:1573–1594
Acknowledgements
We thank the staff of the laboratory for their technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31471844), the PhD Research Startup Foundation of Northwest A&F University (No. Z109021621), and the Central College Basic Scientific Research Business Expenses Special Funds (No. Z109021606).
Author information
Authors and Affiliations
Contributions
DM, YX, and CT collected samples. WK and LR analyzed the transcriptome data. XY, LG, and LZ conceived the study and participated in the design and coordination. LR drafted the manuscript. ZC and LTM modified the language. XG designed primers for qRT-PCR. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Handling Editor: Hanns H. Kassemeyer
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, R., Weng, K., Dou, M. et al. Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola. Protoplasma 256, 1409–1424 (2019). https://doi.org/10.1007/s00709-019-01387-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-019-01387-x