Abstract
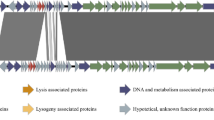
Klebsiella pneumoniae is an important human pathogen that is associated with a wide range of diseases, including pneumonia and septicemia. Because of the threat of drug-resistant K. pneumoniae to humans, especially carbapenem-resistant K. pneumoniae, which is becoming a growing threat to hospitalized patients, the potential use of phage therapy has generated considerable interest. Henu1, isolated from a sewage sample, was identified as a linear double-stranded DNA phage of 40,352 bp with 53.14% G + C content and 143-bp terminal repeats. The Henu1 genome contains 45 open reading frames, and no tRNA genes were found. K. pneumoniae clinical strains with the capsular types K-1, K-2, and K-57 could be infected by Henu1. No human-virulence-related genes or lysogen-formation gene clusters were detected in this phage genome, suggesting that Henu1 is a virulent phage in its bacterial host and is safe for humans.




Similar content being viewed by others
References
Hildebrand A, Mbiegert R (2010) Fast and accurate automatic structure prediction with HHpred. Proteins Struct Funct Bioinform 77:128–132
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kusradze I, Karumidze N, Rigvava S, Dvalidze T, Katsitadze M, Amiranashvili I, Goderdzishvili M (2016) Characterization and testing the efficiency of Acinetobacter baumannii vB-GEC_Ab-M-G7 as an antibacterial agent. Front Microbiol 7:1590
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids Res 25:955–964
Martin RM, Bachman MA (2018) Colonization, Infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 8:4
Pan YJ, Lin TL, Chen CC, Tsai YT, Cheng YH, Chen YY, Hsieh PF, Lin YT, Wang JT (2017) Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types. J Virol 91:JVI.02457-02416
Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603
Schuch R, Khan Mirzaei M, Nilsson AS (2015) Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. Plos One 10:e0118557
Shang A, Liu Y, Wang J, Mo Z, Li G, Mou H (2015) Complete nucleotide sequence of Klebsiella phage P13 and prediction of an EPS depolymerase gene. Virus Genes 50:1–11
Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY (2012) Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887
Tabassum R, Shafique M, Khawaja KA, Alvi IA, Rehman Y, Sheik CS, Abbas Z, Rehman S (2018) Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae. Sci Rep 2018:8
Taha OA, Connerton PL, Connerton IF, El-Shibiny A (2018) Bacteriophage ZCKP1: a potential treatment for Klebsiella pneumoniae isolated from diabetic foot patients. Front Microbiol 9:2127
Volozhantsev NV, Myakinina VP, Popova AV, Kislichkina AA, Komisarova EV, Knyazeva AI, Krasilnikova VM, Fursova NK, Svetoch EA (2015) Complete genome sequence of novel T7-like virus vB_KpnP_KpV289 with lytic activity against Klebsiella pneumoniae. Arch Virol 161:499–501
Funding
The Natural Science Foundation of Education Department of Henan Province under Grant 17A310015.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Teng, T., Li, Q., Liu, Z. et al. Characterization and genome analysis of novel Klebsiella phage Henu1 with lytic activity against clinical strains of Klebsiella pneumoniae. Arch Virol 164, 2389–2393 (2019). https://doi.org/10.1007/s00705-019-04321-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04321-x