Abstract
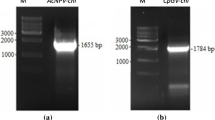
Dendrolimus kikuchii Matsumura nucleopolyhedrovirus (DkNPV) is a novel nucleopolyhedrovirus strain that has exhibited high potential as biological control agent against D. kikuchii. In this work, a 1755-bp DkChi gene with sequence homology to a chitinase gene was cloned from the genomic DNA of DkNPV using a DNA fragment library. The DkChi gene, encoding 558 residues protein with a predicted mass of 61.6 kDa, was expressed at high levels in Escherichia coli and purified by affinity chromatography. We confirmed that the prepared protein was the DkChi protein by mass spectrometry analysis. Enzyme activity analysis showed that DkChi had both endo- and exo-chitinase activities. Interestingly, the DkChi protein displayed a strong insecticidal activity against Spodoptera exigua, Hyphantria cunea, Helicoverpa armigera and Lymantria dispar. The results suggest that DkChi is a good candidate protein for significantly contributing to pest control.





Similar content being viewed by others
References
Kramer KJ, Muthukrishnan S (2005) Chitin metabolism in insects. In: Gilbert LI, Iatrou K, Gill S (eds) Comprehensive molecular insect science. Elsevier, Oxford, pp 111–144
Cohen-Kupiec R, Chet I (1998) The molecular biology of chitin digestion. Curr Opin Biotechnol 9:270–277
Kasprzewska A (2003) Plant chitinases-regulation and function. Cell Mol Biol Lett 8:809–824
Sharma N, Sharma KP, Gaur RK, Gupta VK (2011) Role of chitinase in plant defense. Asian J Biochem 6:29–37
McCreath KJ, Gooday GW (1992) A rapid and sensitive microassay for determination of chitinolytic activity. J Microbiol Methods 14:229–237
Brurberg MB, Synstad B, Klemsdal SS, van Aalten DMF, Sundheim L, Eijsink VGH (2001) Chitinases from Serratia marcescens. Recent Res Dev Microbiol 5:187–204
Tews I, Vincentelli R, Vorgias CE (1996) N-Acetylglucosaminidase (chitobiase) from Serratia marcescens: gene sequence, and protein production and purification in Escherichia coli. Gene 170:63–67
Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilsion KS, Vorgias CE (1996) Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol 3:638–648
Williams SJ, Mark BL, Vocadlo DJ, James MN, Withers SG (2002) Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J Biol Chem 277:40055–40065
Wang H, Wu D, Deng F, Peng H, Chen X, Lauzon H, Arif BM, Jehle JA, Hu Z (2004) Characterization and phylogenetic analysis of the chitinase gene from the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. Virus Res 100:179–189
Hawtin RE, Arnold K, Ayres MD, Zanotto PM, Howard SC, Gooday GA, Chappell LH, Kitts PA, King LA, Possee RD (1995) Identification and preliminary characherization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology 212:673–685
Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, Kuzio JA, Possee RD (1997) Liquefaction of Autographa californica nucleopolyhedrovirus infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243–253
Thomas CJ, Brown HL, Hawes CR, Lee BY, Min M, King LA, Possee RD (1998) Localization of a baculovirus-induced chitinase in the insect cell endoplasmic reticulum. J Virol 72:10207–10212
Saville GP, Thomas CJ, Possee RD, King LA (2002) Partial redistribution of the Autographa californica nucleopolyhedrovirus chitinase in virus-infected cells accompanies mutation of the carboxy-terminal KDEL ER-retention motif. J Gen Virol 83:685–694
Saville GP, Patmanidi AL, Possee RD, King LA (2004) Deletion of the Autographa californica nucleopolyhedrovirus chitinase KDEL motif and in vitro and in vivo analysis of the modified virus. J Gen Virol 85:821–831
Rao R, Fiandra L, Giordana B, de Eguileor M, Congiu T, Burlini N, Arciello S, Corrado G, Pennacchio F (2004) AcMNPV ChiA protein disrupts the peritrophic membrane and alters midgut physiology of Bombyx mori larvae. Insect Biochem Mol Biol 34:1205–1213
Wang F, Zhang C, Shyam Kumar V, Wu X (2005) Influences of chitinase gene deletion from BmNPV on the cell lysis and host liquefaction. Arch Virol 150:981–990
Ohkawa T, Majima K, Maeda S (1994) A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J Virol 68:6619–6625
Hom LG, Volkman LE (2000) Autographa californica M nucleopolyhedrovirus chiA is required for processing of V-CATH. Virology 277:178–183
Slack JM, Kuzio J, Faulkner P (1995) Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa calfornica multiple nuclear polyhedrosis virus. J Gen Virol 76:1091–1098
Yang M, Li M, Wang Y, Qu L, Huai K, Nie X, Qiao L, Ding J, Zhang Y (2011) Virulence and characteristics of a new nucleopolyhedrovirus strain of Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Afr J Microbiol Res 5:2261–2265
Yang D, Wang Y, Li Y (1986) Studies on the nuclear polyhedrosis virus disease of simao pine caterpil-lcar infected by Dendrolimus kikuchii Matsumura (Lepidoptera: Lasiocampidae). J Yunnan Univ 1:105–110
Rajamohan F, Cotrill JA, Gould F, Dean DH (1996) Role of domain II, loop 2 residues of Bacillus thuringiensis CryIAb delta-endotoxin in reversible and irreversible binding to Manduca sexta and Heliothis virescens. J Biol Chem 271:2390–2396
Finney DJ (1971) Probit analysis, 3rd edn. Cambridge University Press, Cambridge
Uchiyama T, Katouno F, Nikaidou N, Nonaka T, Sugiyama J, Watanabe T (2001) Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J Biol Chem 276:41343–41349
Young VL, Simpson RM, Ward VK (2005) Characterization of an exochitinse from Epiphyas postvittana nucleopolyhedrovirus (family Baculoviridae). J Gen Virol 86:3253–3261
Gomi S, Majima K, Maeda S (1999) Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol 80:1323–1337
Perrakis A, Tews I, Dauter Z, Oppenheim AB, Chet I, Wilson KS, Vorgias CE (1994) Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169–1180
Bork P, Doolittle RF (1992) Proposed acquisition of an animal protein domain by bacteria. Proc Natl Acad Sci USA 89:8990–8994
Perrakis A, Ouzounis C, Wilson KS (1997) Evolution of immunoglobulin-like modules in chitinases: their structural flexibility and functional implications. Fold Des 2:291–294
Acknowledgments
This work was supported by the Program of National Nature Science Foundation of China (grant no. 31200491) and State 863 Project funded by Ministry of Science and Technology (grant no. 2012AA101503), P.R. China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2013_1775_MOESM1_ESM.tif
Fig. S1 Phylogenetic tree of chitinases from baculovirus. The tree was constructed by the neighbor-joining method, and bootstrap values are indicated at the branches. The amino acid sequences of chitinases were from the following viruses: Dendrolimus kikuchii NPV (JN680874), Hyphantria cunea NPV (YP473218), Orgyia pseudotsugata MNPV (NP046280), Bombyx mori NPV (NP047523), Autographa californica MNPV (NP054156), Spodoptera exigua MNPV (NP037779), Helicoverpa armigera SNPV (AAK28347) and Spodoptera litura NPV (NP258310) (TIFF 292 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Qu, L., Zhang, Z. et al. Characterization of a novel chitinase, DkChi, from Dendrolimus kikuchii nucleopolyhedrovirus. Arch Virol 158, 2523–2530 (2013). https://doi.org/10.1007/s00705-013-1775-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1775-7