Abstract
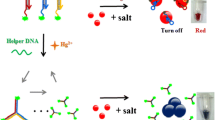
An electrochemiluminescence (ECL) based method is described for the determination of the activity of the enzyme uracil-DNA glycosylase (UDG). It is based on the use of nicking enzyme-assisted signal amplification and catalytic hairpin assembly. UDG can recognize and hydrolyze the uracil bases from the stem of hairpin DNA1 (HP1). This causes the opening of HP1 to form a straight strand DNA. The straight HP1 can hybridize with hairpin DNA2 (HP2) to form a DNA duplex. In the presence of nicking enzyme, it can recognize and cut the specific sequences in the HP2 of the DNA duplex, and a subsequent release of HP1. It hybridizes with other HP2 to trigger the continuous cleavage of HP2, concomitantly generating abundant intermediate sequences (S1). The hairpin DNA3 (HP3) is immobilized on a gold electrode via Au-S chemistry. In the presence of S1, HP3 hybridizes with S1 and its hairpin structure is opened. This hybridization causes displacement from hairpin DNA4 (HP4), and S1 is released to initiate the next hybridization process. Thus, a massive number of HP3-HP4 duplexes is generated after the cyclic process. Subsequently, the cDNA modified on bio-bar-coded AuNP-CdSe quantum dots are immobilized on the electrode by hybridization with the redundant part of the opened HP4. This results in a significant amplification of the ECL signal. This biosensor is sensitive and selective for UDG. The detection limit is 6 mU·mL−1 and the dynamic range extends from 0.02 to 22 U·mL−1. The method was applied to real samples and gained good performance, thereby providing an ideal way for DNA repair enzyme-related biomedical research and diagnosis.

Schematic presentation of the electrochemiluminescence (ECL) detection of uracil-DNA glycosylase (UDG) based on nicking enzyme assisted signal amplification and catalyzed hairpin assembly. The bio-barcoded Au NP-CdSe QDs serve as the ECL signal probes to achieve a significantly signal amplification.





Similar content being viewed by others
References
van Loon AAWM, Sonneveld E, Hoogerbrugge J, van der Schans GP, Grootegoed JA, Lohman PHM, Baan RA (1993) Induction and repair of D N A single-strand breaks and D N A base damage at different cellular stages of spermatogenesis of the hamster upon in vitro exposure to ionizing radiation. Mutat Res, DNA Repair 294:139–148
Giovannelli L, Pitozzi V, Moretti S, Boddi V, Dolara P (2006) Seasonal variations of DNA damage in human lymphocytes: correlation with different environmental variables. Mutat Res 593:143–152
Papachristou F, Simopoulou M, Touloupidis S, Tsalikidis C, Sofikitis N, Lialiaris T (2008) DNA damage and chromosomal aberrations in various types of male factor infertility. Fertil Steril 90:1774–1781
Krokan HE, Drabløs F, Slupphaug G (2002) Uracil in DNA – occurrence, consequences and repair. Oncogene 21:8935–8948
Krokan HE, Standal R, Slupphaug G (1997) DNA glycosylases in the base excision repair of DNA. Biochem J 325:1–16
Impellizzeri KJ, Anderson B, Burgers PMJ (1991) The Spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J Bacteriol 173:6807–6810
Lindahl T (1974) An N-Glycosidase from Escherichia coli that releases free uracil from DNA containing Deaminated cytosine residues. Proc Natl Acad Sci U S A 71:3649–3653
Wallace SS (2014) Base excision repair: a critical player in many games. DNA Repair 19:14–26
Duncan BK, Miller JH (1980) Mutagetic deamination of cytosine residues in DNA. Nature 287:560–561
Wang Y, Wang Y, Li D, Xu J, Ye C (2018) Detection of nucleic acids and elimination of carryover contamination by using loop-mediated isothermal amplification and antarctic thermal sensitive uracil-DNA-glycosylase in a lateral flow biosensor: application to the detection of Streptococcus pneumonia. Microchim Acta 185:212
Wu YS, Yan P, Xu XW, Jiang W (2016) A unique dual recognition hairpin probe mediated fluorescent amplification method for sensitive detection of uracil-DNA glycosylase and endonuclease IV activities. Analyst 141:1789–1795
Sousa MML, Krokan HE, Slupphaug G (2007) DNA-uracil and human pathology. Mol Asp Med 28:276–306
Sung JS, Mosbaugh DW (2000) Escherichia coli double-Strand uracil-DNA glycosylase: involvement in uracil-mediated DNA Base excision repair and stimulation of activity by endonuclease IV. Biochemistry 39:10224–10235
Duthie SJ, McMillan P (1997) Uracil misincorporation in human DNA detected using single cell gel electrophoresis. Carcinogenesis 18:1709–1714
Krokan H, Wittwer CU (1981) Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res 9:2599–2613
Wang LJ, Ren M, Zhang QY, Tang B, Zhang CY (2017) Excision repair-initiated enzyme-assisted bicyclic Cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 89:4488–4494
Richter MM (2004) Electrochemiluminescence (ECL). Chem Rev 104:3003–3036
Shi CG, Shan X, Pan ZQ, Xu JJ, Lu C, Bao N, Gu HY (2012) Quantum Dot (QD)-modified carbon tape electrodes for reproducible Electrochemiluminescence (ECL) emission on a PaperBased platform. Anal Chem 84:3033–3038
Liu Q, Ma C, Liu XP, Wei YP, Mao CJ, Zhu JJ (2017) A novel electrochemiluminescence biosensor for the detection of microRNAs based on a DNA functionalized nitrogen doped carbon quantum dots as signal enhancers. Biosens Bioelectron 92:273–279
Wang MH, Zhou YL, YL YHS, Jiang WJ, Wang HY, Ai SY (2018) Signal-on electrochemiluminescence biosensor for microRNA-319a detection based on two-stage isothermal strand-displacement polymerase reaction. Biosens Bioelectron 107:34–39
Liu Q, Yang Y, Liu XP, Wei YP, Mao CJ, Chen JS, Niu HL, Song JM, Zhang SY, Jin BK, Jiang M (2017) A facile in situ synthesis of MIL-101-CdSe nanocomposites for ultrasensitive electrochemiluminescence detection of carcinoembryonic antigen. Sensors Actuators B Chem 242:1073–1078
Hu XW, Mao CJ, Song JM, Niu HL, Zhang SY, Huang HP (2013) Fabrication of GO/PANi/CdSe nanocomposites for sensitive electrochemiluminescence biosensor. Biosens Bioelectron 41:372–378
Cheng CM, Huang Y, Tian XQ, Zheng BZ, Li Y, Yuan HY, Xiao D, Xie SP, Choi MMF (2012) Electrogenerated Chemiluminescence behavior of graphite-like carbon nitride and its application in selective sensing Cu2+. Anal Chem 84:4754–4759
Zhou ZX, Shang QW, Shen YF, Zhang LQ, Zhang YY, Lv YQ, Li Y, Liu SQ, Zhang YJ (2016) Chemically modulated carbon nitride Nanosheets for highly selective Electrochemiluminescent detection of multiple metal-ions. Anal Chem 88:6004–6010
Chan SH, Stoddard BL, Xu SY (2011) Natural and engineered nicking endonucleases-from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res 39:11–18
Kuhn H, Kamenetskii MDF (2008) Labeling of unique sequences in double-stranded DNA at sites of vicinal nicks generated by nicking endonucleases. Nucleic Acids Res 36:e40
Bi S, Zhang JL, Zhang SS (2010) Ultrasensitive and selective DNA detection based on nicking endonuclease assisted signal amplification and its application in cancer cell detection. Chem Commun 46:5509–5511
Zhu XL, Zhao J, Wu Y, Shen ZM, Li GX (2011) Fabrication of a highly sensitive Aptasensor for potassium with a nicking endonuclease-assisted signal amplification strategy. Anal Chem 83:4085–4089
Liu XJ, Chen MQ, Hou T, Wang XZ, Liu SF, Li F (2014) Label-free colorimetric assay for base excision repair enzyme activity based on nicking enzyme assisted signal amplification. Biosens Bioelectron 54:598–602
Zhang PP, Wang L, Zhao HY, Xua XW, Jia W (2017) Self-primer and self-template recycle rolling circle amplification strategy for sensitive detection of uracil-DNA glycosylase activity. Anal Chim Acta 1001:119–124
Kong XJ, Wu S, Cen Y, Yu RQ, Chu X (2016) “Light-up” sensing of human 8-oxoguanine DNA glycosylase activity by target-induced autocatalytic DNAzyme-generated rolling circle amplification. Biosens Bioelectron 79:679–684
Wu YS, Wang L, Jiang W (2016) Toehold-mediated strand displacement reaction-dependent fluorescent strategy for sensitive detection of uracil-DNA glycosylase activity. Biosens Bioelectron 89:984–988
Zheng AX, Li J, Wang JR, Song XR, Chen GN, Yan HH (2012) Enzyme-free signal amplification in the DNAzyme sensor via target-catalyzed hairpin assembly. Chem Commun 48:3112–3114
Li YL, Yu C, Yang B, Liu ZR, Xia PY, Wang Q (2018) Target-catalyzed hairpin assembly and metal-organic frameworks mediated nonenzymatic co-reaction for multiple signal amplification detection of miR-122 in human serum. Biosens Bioelectron 102:307–315
Wu XY, Chai YQ, YQ ZP, Yuan R (2015) An electrochemical biosensor for sensitive detection of MicroRNA-155: combining target recycling with Cascade catalysis for signal amplification. ACS Appl Mater Interfaces 7:713–720
Liu SF, Wang Y, Ming JJ, Lin Y, Cheng CB, Li F (2013) Enzyme-free and ultrasensitive electrochemical detection of nucleic acids by target catalyzed hairpin assembly followed with hybridization chain reaction. Biosens Bioelectron 49:472–477
Lin DJ, Wu J, Yan F, Deng SY, Ju HX (2011) Ultrasensitive immunoassay of protein biomarker based on Electrochemiluminescent quenching of quantum dots by hemin bio-Bar-coded nanoparticle tags. Anal Chem 83:5214–5221
Liu XL, Ma CC, Yan Y, Yao GX, Tang YF, Huo PW, Shi WD, Yan YS (2013) Hydrothermal synthesis of CdSe quantum dots and their photocatalytic activity on degradation of Cefalexin. Ind Eng Chem Res 52:15015–15023
Nie HJ, Wang W, Li W, Nie Z, Yao SZ (2015) A colorimetric and smartphone readable method for uracil-DNA glycosylase detection based on the target-triggered formation of G-quadruplex. Analyst 140:2771–2777
Zhang XF, Li N, Ling Y, Tang L, Li NB, Luo HQ (2018) Linked bridge hybridizing-induced split G quadruplex DNA machine and its application to uracil-DNA glycosylase detection. Sensors Actuators B Chem 255:2589–2594
Lee CY, Parka KS, Park HG (2015) Fluorescent G-quadruplex probe for the assay of base excision repair enzyme activity. Chem Commun 51:13744–13747
Acknowledgements
We are grateful to Natural Science Research Key Project of Education Department of Anhui Province (No. KJ2018A0453, KJ2017A434, KJ2017A435 and KJ2018A0446) and Academic Technical Leader of Suzhou University (No. 2016xjxs03 and 2016msgzs071) for funding this work. The authors also thank the Key Discipline of Material Science and Engineering of Suzhou University (No. 2017XJZDXK3) and Innovative Research Team of Anhui Provincial Education Department (No. 2016SCXPTTD) for supporting this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 388 kb)
Rights and permissions
About this article
Cite this article
Liu, Q., Liu, C., Zhu, G. et al. Electrochemiluminescent determination of the activity of uracil-DNA glycosylase: Combining nicking enzyme assisted signal amplification and catalyzed hairpin assembly. Microchim Acta 186, 179 (2019). https://doi.org/10.1007/s00604-019-3280-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3280-5