Abstract
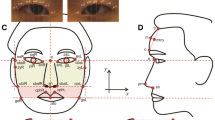
In human society, the facial surface is visible and recognizable based on the facial shape variation which represents a set of highly polygenic and correlated complex traits. Understanding the genetic basis underlying facial shape traits has important implications in population genetics, developmental biology, and forensic science. A number of single nucleotide polymorphisms (SNPs) are associated with human facial shape variation, mostly in European populations. To bridge the gap between European and Asian populations in term of the genetic basis of facial shape variation, we examined the effect of these SNPs in a European–Asian admixed Eurasian population which included a total of 612 individuals. The coordinates of 17 facial landmarks were derived from high resolution 3dMD facial images, and 136 Euclidean distances between all pairs of landmarks were quantitatively derived. DNA samples were genotyped using the Illumina Infinium Global Screening Array and imputed using the 1000 Genomes reference panel. Genetic association between 125 previously reported facial shape-associated SNPs and 136 facial shape phenotypes was tested using linear regression. As a result, a total of eight SNPs from different loci demonstrated significant association with one or more facial shape traits after adjusting for multiple testing (significance threshold p < 1.28 × 10−3), together explaining up to 6.47% of sex-, age-, and BMI-adjusted facial phenotype variance. These included EDAR rs3827760, LYPLAL1 rs5781117, PRDM16 rs4648379, PAX3 rs7559271, DKK1 rs1194708, TNFSF12 rs80067372, CACNA2D3 rs56063440, and SUPT3H rs227833. Notably, the EDAR rs3827760 and LYPLAL1 rs5781117 SNPs displayed significant association with eight and seven facial phenotypes, respectively (2.39 × 10−5 < p < 1.28 × 10−3). The majority of these SNPs showed a distinct allele frequency between European and East Asian reference panels from the 1000 Genomes Project. These results showed the details of above eight genes influence facial shape variation in a Eurasian population.


Similar content being viewed by others
Change history
30 August 2021
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s00439-021-02352-6
References
Adhikari K, Reales G, Smith AJ, Konka E, Palmen J, Quinto-Sanchez M, Acuna-Alonzo V, Jaramillo C, Arias W, Fuentes M, Pizarro M, Barquera Lozano R, Macin Perez G, Gomez-Valdes J, Villamil-Ramirez H, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Hurtado M, Villegas V, Granja V, Gallo C, Poletti G, Schuler-Faccini L, Salzano FM, Bortolini MC, Canizales-Quinteros S, Rothhammer F, Bedoya G, Calderon R, Rosique J, Cheeseman M, Bhutta MF, Humphries SE, Gonzalez-Jose R, Headon D, Balding D, Ruiz-Linares A (2015) A genome-wide association study identifies multiple loci for variation in human ear morphology. Nat Commun 6:7500. https://doi.org/10.1038/ncomms8500
Adhikari K, Fuentes-Guajardo M, Quinto-Sanchez M, Mendoza-Revilla J, Camilo Chacon-Duque J, Acuna-Alonzo V, Jaramillo C, Arias W, Lozano RB, Perez GM, Gomez-Valdes J, Villamil-Ramirez H, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Hurtado M, Villegas V, Granja V, Gallo C, Poletti G, Schuler-Faccini L, Salzano FM, Bortolini MC, Canizales-Quinteros S, Cheeseman M, Rosique J, Bedoya G, Rothhammer F, Headon D, Gonzalez-Jose R, Balding D, Ruiz-Linares A (2016) A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat Commun 7:11616. https://doi.org/10.1038/ncomms11616
Alkhudhairi TD, Alkofide EA (2010) Cephalometric craniofacial features in Saudi parents and their offspring. Angle Orthod 80:1010–1017. https://doi.org/10.2319/050410-66.1
Bryk J, Hardouin E, Pugach I, Hughes D, Strotmann R, Stoneking M, Myles S (2008) Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS One 3:e2209. https://doi.org/10.1371/journal.pone.0002209
Cha S, Lim JE, Park AY, Do JH, Lee SW, Shin C, Cho NH, Kang JO, Nam JM, Kim JS, Woo KM, Lee SH, Kim JY, Oh B (2018) Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics 19:481. https://doi.org/10.1186/s12864-018-4865-9
Claes P, Roosenboom J, White JD, Swigut T, Sero D, Li J, Lee MK, Zaidi A, Mattern BC, Liebowitz C, Pearson L, Gonzalez T, Leslie EJ, Carlson JC, Orlova E, Suetens P, Vandermeulen D, Feingold E, Marazita ML, Shaffer JR, Wysocka J, Shriver MD, Weinberg SM (2018) Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat Genet 50:414–423. https://doi.org/10.1038/s41588-018-0057-4
Cole JB, Manyama M, Kimwaga E, Mathayo J, Larson JR, Liberton DK, Lukowiak K, Ferrara TM, Riccardi SL, Li M, Mio W, Prochazkova M, Williams T, Li H, Jones KL, Klein OD, Santorico SA, Hallgrimsson B, Spritz RA (2016) Genomewide association study of African children identifies association of SCHIP1 and PDE8A with facial size and shape. PLoS Genet 12:e1006174. https://doi.org/10.1371/journal.pgen.1006174
Cole JB, Manyama M, Larson JR, Liberton DK, Ferrara TM, Riccardi SL, Li M, Mio W, Klein OD, Santorico SA, Hallgrimsson B, Spritz RA (2017) Human facial shape and size heritability and genetic correlations. Genetics 205:967–978. https://doi.org/10.1534/genetics.116.193185
Crouch DJM, Winney B, Koppen WP, Christmas WJ, Hutnik K, Day T, Meena D, Boumertit A, Hysi P, Nessa A, Spector TD, Kittler J, Bodmer WF (2018) Genetics of the human face: identification of large-effect single gene variants. Proc Natl Acad Sci USA 115:E676–E685. https://doi.org/10.1073/pnas.1708207114
Delaneau O, Zagury JF, Marchini J (2013) Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10:5–6. https://doi.org/10.1038/nmeth.2307
Fujimoto A, Kimura R, Ohashi J, Omi K, Yuliwulandari R, Batubara L, Mustofa MS, Samakkarn U, Settheetham-Ishida W, Ishida T, Morishita Y, Furusawa T, Nakazawa M, Ohtsuka R, Tokunaga K (2008a) A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Hum Mol Genet 17:835–843. https://doi.org/10.1093/hmg/ddm355
Fujimoto A, Ohashi J, Nishida N, Miyagawa T, Morishita Y, Tsunoda T, Kimura R, Tokunaga K (2008b) A replication study confirmed the EDAR gene to be a major contributor to population differentiation regarding head hair thickness in Asia. Hum Genet 124:179–185. https://doi.org/10.1007/s00439-008-0537-1
Gene Ontology C (2015) Gene ontology consortium: going forward. Nucleic Acids Res 43:D1049–D1056. https://doi.org/10.1093/nar/gku1179
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. https://doi.org/10.1038/nature11632
Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, Zuk O, Lander ES, Schaffner SF, Sabeti PC (2010) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327:883–886. https://doi.org/10.1126/science.1183863
Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, Cabili M, Adegbola RA, Bamezai RN, Hill AV, Vannberg FO, Rinn JL, Genomes P, Lander ES, Schaffner SF, Sabeti PC (2013) Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. https://doi.org/10.1016/j.cell.2013.01.035
Guo J, Mei X, Tang K (2013) Automatic landmark annotation and dense correspondence registration for 3D human facial images. BMC Bioinformatics 14:232. https://doi.org/10.1186/1471-2105-14-232
Hartigan JA, Wong MA (1979) Algorithm AS 136: a K-means clustering algorithm. J R Stat Soc 28:100–108
Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Lango Allen H, Weyant RJ, Wheeler E, Wood AR, Magic Estrada K, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K et al (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 42:949–960. https://doi.org/10.1038/ng.685
Hotta K, Kitamoto A, Kitamoto T, Mizusawa S, Teranishi H, So R, Matsuo T, Nakata Y, Hyogo H, Ochi H, Nakamura T, Kamohara S, Miyatake N, Kotani K, Itoh N, Mineo I, Wada J, Yoneda M, Nakajima A, Funahashi T, Miyazaki S, Tokunaga K, Masuzaki H, Ueno T, Chayama K, Hamaguchi K, Yamada K, Hanafusa T, Oikawa S, Sakata T, Tanaka K, Matsuzawa Y, Nakao K, Sekine A (2013) Replication study of 15 recently published Loci for body fat distribution in the Japanese population. J Atheroscler Thromb 20:336–350
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5:e1000529. https://doi.org/10.1371/journal.pgen.1000529
Howie B, Marchini J, Stephens M (2011) Genotype imputation with thousands of genomes. G3 (Bethesda) 1:457–470. https://doi.org/10.1534/g3.111.001198
Kamberov YG, Wang S, Tan J, Gerbault P, Wark A, Tan L, Yang Y, Li S, Tang K, Chen H, Powell A, Itan Y, Fuller D, Lohmueller J, Mao J, Schachar A, Paymer M, Hostetter E, Byrne E, Burnett M, McMahon AP, Thomas MG, Lieberman DE, Jin L, Tabin CJ, Morgan BA, Sabeti PC (2013) Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 152:691–702. https://doi.org/10.1016/j.cell.2013.01.016
Kimura R, Yamaguchi T, Takeda M, Kondo O, Toma T, Haneji K, Hanihara T, Matsukusa H, Kawamura S, Maki K, Osawa M, Ishida H, Oota H (2009) A common variation in EDAR is a genetic determinant of shovel-shaped incisors. Am J Hum Genet 85:528–535. https://doi.org/10.1016/j.ajhg.2009.09.006
Lee MK, Shaffer JR, Leslie EJ, Orlova E, Carlson JC, Feingold E, Marazita ML, Weinberg SM (2017a) Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. Plos One. https://doi.org/10.1371/journal.pone.0176566(ARTN e0176566)
Lee MK, Shaffer JR, Leslie EJ, Orlova E, Carlson JC, Feingold E, Marazita ML, Weinberg SM (2017b) Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One 12:e0176566. https://doi.org/10.1371/journal.pone.0176566
Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95:221–227. https://doi.org/10.1038/sj.hdy.6800717
Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, Speliotes EK, Thorleifsson G, Willer CJ, Herrera BM, Jackson AU, Lim N, Scheet P, Soranzo N, Amin N, Aulchenko YS, Chambers JC, Drong A, Luan J, Lyon HN, Rivadeneira F, Sanna S, Timpson NJ, Zillikens MC, Zhao JH, Almgren P, Bandinelli S, Bennett AJ, Bergman RN, Bonnycastle LL, Bumpstead SJ, Chanock SJ, Cherkas L, Chines P, Coin L, Cooper C, Crawford G, Doering A, Dominiczak A, Doney AS, Ebrahim S, Elliott P, Erdos MR, Estrada K, Ferrucci L, Fischer G, Forouhi NG, Gieger C, Grallert H, Groves CJ, Grundy S, Guiducci C, Hadley D, Hamsten A, Havulinna AS, Hofman A, Holle R, Holloway JW, Illig T, Isomaa B, Jacobs LC, Jameson K, Jousilahti P, Karpe F, Kuusisto J, Laitinen J, Lathrop GM, Lawlor DA, Mangino M, McArdle WL, Meitinger T, Morken MA, Morris AP, Munroe P, Narisu N, Nordstrom A, Nordstrom P, Oostra BA, Palmer CN, Payne F, Peden JF, Prokopenko I, Renstrom F, Ruokonen A, Salomaa V, Sandhu MS, Scott LJ, Scuteri A, Silander K, Song K, Yuan X, Stringham HM, Swift AJ, Tuomi T, Uda M, Vollenweider P, Waeber G, Wallace C, Walters GB, Weedon MN et al (2009) Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet 5:e1000508. https://doi.org/10.1371/journal.pgen.1000508
Liu F, van der Lijn F, Schurmann C, Zhu G, Chakravarty MM, Hysi PG, Wollstein A, Lao O, de Bruijne M, Ikram MA, van der Lugt A, Rivadeneira F, Uitterlinden AG, Hofman A, Niessen WJ, Homuth G, de Zubicaray G, McMahon KL, Thompson PM, Daboul A, Puls R, Hegenscheid K, Bevan L, Pausova Z, Medland SE, Montgomery GW, Wright MJ, Wicking C, Boehringer S, Spector TD, Paus T, Martin NG, Biffar R, Kayser M (2012) A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet 8:e1002932. https://doi.org/10.1371/journal.pgen.1002932
Liu CT, Buchkovich ML, Winkler TW, Heid IM, African Ancestry Anthropometry Genetics C, Consortium G, Borecki IB, Fox CS, Mohlke KL, North KE, Adrienne Cupples L (2014) Multi-ethnic fine-mapping of 14 central adiposity loci. Hum Mol Genet 23:4738–4744. https://doi.org/10.1093/hmg/ddu183
Lv D, Zhou D, Zhang Y, Zhang S, Zhu YM (2017) Two obesity susceptibility loci in LYPLAL1 and ETV5 independently associated with childhood hypertension in Chinese population. Gene 627:284–289. https://doi.org/10.1016/j.gene.2017.06.030
Members BIGDC (2018) Database resources of the BIG data center in 2018. Nucleic Acids Res 46:D14–D20. https://doi.org/10.1093/nar/gkx897
Mou C, Thomason HA, Willan PM, Clowes C, Harris WE, Drew CF, Dixon J, Dixon MJ, Headon DJ (2008) Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum Mutat 29:1405–1411. https://doi.org/10.1002/humu.20795
Nettleton JA, Follis JL, Ngwa JS, Smith CE, Ahmad S, Tanaka T, Wojczynski MK, Voortman T, Lemaitre RN, Kristiansson K, Nuotio ML, Houston DK, Perala MM, Qi Q, Sonestedt E, Manichaikul A, Kanoni S, Ganna A, Mikkila V, North KE, Siscovick DS, Harald K, McKeown NM, Johansson I, Rissanen H, Liu Y, Lahti J, Hu FB, Bandinelli S, Rukh G, Rich S, Booij L, Dmitriou M, Ax E, Raitakari O, Mukamal K, Mannisto S, Hallmans G, Jula A, Ericson U, Jacobs DR Jr, Van Rooij FJ, Deloukas P, Sjogren P, Kahonen M, Djousse L, Perola M, Barroso I, Hofman A, Stirrups K, Viikari J, Uitterlinden AG, Kalafati IP, Franco OH, Mozaffarian D, Salomaa V, Borecki IB, Knekt P, Kritchevsky SB, Eriksson JG, Dedoussis GV, Qi L, Ferrucci L, Orho-Melander M, Zillikens MC, Ingelsson E, Lehtimaki T, Renstrom F, Cupples LA, Loos RJ, Franks PW (2015) Gene x dietary pattern interactions in obesity: analysis of up to 68 317 adults of European ancestry. Hum Mol Genet 24:4728–4738. https://doi.org/10.1093/hmg/ddv186
Park JH, Yamaguchi T, Watanabe C, Kawaguchi A, Haneji K, Takeda M, Kim YI, Tomoyasu Y, Watanabe M, Oota H, Hanihara T, Ishida H, Maki K, Park SB, Kimura R (2012) Effects of an Asian-specific nonsynonymous EDAR variant on multiple dental traits. J Hum Genet 57:508–514. https://doi.org/10.1038/jhg.2012.60
Paternoster L, Zhurov AI, Toma AM, Kemp JP, St Pourcain B, Timpson NJ, McMahon G, McArdle W, Ring SM, Smith GD, Richmond S, Evans DM (2012) Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am J Hum Genet 90:478–485. https://doi.org/10.1016/j.ajhg.2011.12.021
Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA (2016a) Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 48:709–717. https://doi.org/10.1038/ng.3570
Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA (2016b) Detection and interpretation of shared genetic influences on 42 human traits (vol 48, pg 709, 2016). Nat Genet 48:1296
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, Schaffner SF, Lander ES, International HapMap C, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, et al. (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. https://doi.org/10.1038/nature06250
Shaffer JR, Li J, Lee MK, Roosenboom J, Orlova E, Adhikari K, andMe Research T, Gallo C, Poletti G, Schuler-Faccini L, Bortolini MC, Canizales-Quinteros S, Rothhammer F, Bedoya G, Gonzalez-Jose R, Pfeffer PE, Wollenschlaeger CA, Hecht JT, Wehby GL, Moreno LM, Ding A, Jin L, Yang Y, Carlson JC, Leslie EJ, Feingold E, Marazita ML, Hinds DA, Cox TC, Wang S, Ruiz-Linares A, Weinberg SM (2017) Multiethnic GWAS reveals polygenic architecture of earlobe attachment. Am J Hum Genet 101:913–924. https://doi.org/10.1016/j.ajhg.2017.10.001
Shaffer JR, Orlova E, Lee MK, Leslie EJ, Raffensperger ZD, Heike CL, Cunningham ML, Hecht JT, Kau CH, Nidey NL, Moreno LM, Wehby GL, Murray JC, Laurie CA, Laurie CC, Cole J, Ferrara T, Santorico S, Klein O, Mio W, Feingold E, Hallgrimsson B, Spritz RA, Marazita ML, Weinberg SM (2016) Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genet 12:e1006149. https://doi.org/10.1371/journal.pgen.1006149
Song S, Tian D, Li C, Tang B, Dong L, Xiao J, Bao Y, Zhao W, He H, Zhang Z (2018) Genome variation map: a data repository of genome variations in BIG data center. Nucleic Acids Res 46:D944–D949. https://doi.org/10.1093/nar/gkx986
Tan J, Yang Y, Tang K, Sabeti PC, Jin L, Wang S (2013) The adaptive variant EDARV370A is associated with straight hair in East Asians. Hum Genet 132:1187–1191. https://doi.org/10.1007/s00439-013-1324-1
Wang T, Ma X, Peng D, Zhang R, Sun X, Chen M, Yan J, Wang S, Yan D, He Z, Jiang F, Bao Y, Hu C, Jia W (2016) Effects of obesity related genetic variations on visceral and subcutaneous fat distribution in a Chinese population. Sci Rep 6:20691. https://doi.org/10.1038/srep20691
Wroe S, Parr WCH, Ledogar JA, Bourke J, Evans SP, Fiorenza L, Benazzi S, Hublin JJ, Stringer C, Kullmer O, Curry M, Rae TC, Yokley TR (2018) Computer simulations show that Neanderthal facial morphology represents adaptation to cold and high energy demands, but not heavy biting. Proc Biol Sci. https://doi.org/10.1098/rspb.2018.0085
Zaidi AA, Mattern BC, Claes P, McEvoy B, Hughes C, Shriver MD (2017) Investigating the case of human nose shape and climate adaptation. PLoS Genet 13:e1006616. https://doi.org/10.1371/journal.pgen.1006616
Acknowledgements
This project was supported by the National Key R&D Program of China (2017YFC0803501), National Natural Science Foundation of China (91651507), Fund from institute of forensic science (2018JB046), and open projects of the National Engineering Laboratory for Forensic Science (2017NELKFKT05). Biological samples were provided by the National Science and Technological Resources Platform (YCZYPT[2017]01-3 and 2017JB025). Author FL is supported by “The Thousand Talents Plan for Young Professionals”. Author CXL is supported by “The Beijing Leading Talent Program (Z18110006318006)”.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest regarding this work.
Data Availability Statement
The dataset analyzed during the current study is restricted due to participant confidentially. Restrictions apply to the availability of these data, which were used under license for the current study, and so are not entirely publicly available. However, the variation data reported in this paper have been deposited in the Genome Variation Map (Song et al. 2018) in the BIG Data Center (Members BIGDC 2018), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number GVM000031 and can be publicly accessed at http://bigd.big.ac.cn/gvm/getProjectDetail?project=GVM000031.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s00439-021-02352-6
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2019_2023_MOESM2_ESM.tif
Supplementary Figure 2: (A) The relative proportion of variance of the first 20 PCs derived from genotyped data, including our 612 Eurasian samples and the EAS and EUR samples from the 1000 Genomes Project. (B) The PC1 and PC2 plots of genotyped data, including our 612 Eurasian samples and the EAS and EUR samples from the 1000 Genomes Project (TIFF 22719 kb)
Supplementary Figure 3: The hierarchical clustering results of 136 facial phenotypes (TIFF 13680 kb)
439_2019_2023_MOESM4_ESM.tif
Supplementary Figure 4: Worldwide allele-frequency distributions for the eight identified variants. Map displaying the geospatial distribution of the allele frequency of rs3827760 (EDAR), rs5781117 (LYPLAL1), rs4648379 (PRDM16), rs7559271 (PAX3), rs1194708 (DKK1), rs80067372 (TNFSF12), rs56063440 (CACNA2D3), and rs227833 (SUPT3H) across the world. The map is drawn based on allele frequencies of 2504 subjects obtained from 1000 Genome project datasets for each SNP. The pie denotes the sampling locations (TIFF 34735 kb)
About this article
Cite this article
Li, Y., Zhao, W., Li, D. et al. RETRACTED ARTICLE: EDAR, LYPLAL1, PRDM16, PAX3, DKK1, TNFSF12, CACNA2D3, and SUPT3H gene variants influence facial morphology in a Eurasian population. Hum Genet 138, 681–689 (2019). https://doi.org/10.1007/s00439-019-02023-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-02023-7