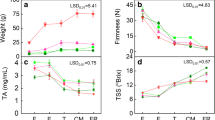
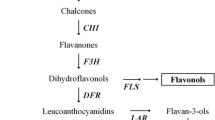
Abstract
Skin color is an important factor in pear breeding programs. The degree of red coloration is determined by the content and composition of anthocyanins. In plants, many MYB transcriptional factors are involved in regulating anthocyanin biosynthesis. In this study, a R2R3-MYB transcription factor gene, PyMYB10, was isolated from Asian pear (Pyrus pyrifolia) cv. ‘Aoguan’. Sequence analysis suggested that the PyMYB10 gene was an ortholog of MdMYB10 gene, which regulates anthocyanin biosynthesis in red fleshed apple (Malus × domestica) cv. ‘Red Field’. PyMYB10 was identified at the genomic level and had three exons, with its upstream sequence containing core sequences of cis-acting regulatory elements involved in light responsiveness. Fruit bagging showed that light could induce expression of PyMYB10 and anthocyanin biosynthesis. Quantitative real-time PCR revealed that PyMYB10 was predominantly expressed in pear skins, buds, and young leaves, and the level of transcription in buds was higher than in skin and young leaves. In ripening fruits, the transcription of PyMYB10 in the skin was positively correlated with genes in the anthocyanin pathway and with anthocyanin biosynthesis. In addition, the transcription of PyMYB10 and genes of anthocyanin biosynthesis were more abundant in red-skinned pear cultivars compared to blushed cultivars. Transgenic Arabidopsis plants overexpressing PyMYB10 exhibited ectopic pigmentation in immature seeds. The study suggested that PyMYB10 plays a role in regulating anthocyanin biosynthesis and the overexpression of PyMYB10 was sufficient to induce anthocyanin accumulation.








Similar content being viewed by others
Abbreviations
- ANS:
-
Anthocyanidin synthase
- AN2:
-
Anthocyanin 2
- BHLH:
-
Basic helix loop helix
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DAFB:
-
Days after bag removed
- DFR:
-
Dihydroflavonol 4-reductase
- F3H:
-
Flavanone 3-hydroxylase
- PAL:
-
Phenylalanine ammonialyase
- SE:
-
Standard error
- Rc:
-
Anti-eukaryotic aspartyl protease
- DEPC:
-
Diethylpyrocarbonate
- C T :
-
Threshold cycle
- ABA:
-
Acrylonitrile butadiene acrylate
- MeJA:
-
Methyl jasmonate
References
Aharoni A, Ric De CH, Wein M, Sun ZK, Greco R, Kroon A, Mol JNM, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28:319–332
Borovsky Y, Oren SM, Ovadia R, De JW, Paran I (2004) The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin 2 of Petunia. Theor Appl Genet 109:23–29
Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7:202–209
Cheng S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dayton DF (1966) The pattern and inheritance of anthocyanin distribution in red pears. J Proc Am Soc Hortic Sci 89:110–116
De Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11:1422–1434
Dussi MC, Sugar D, Wrolstad RE (1995) Characterizing and quantifying anthocyanins in red pears and effect of light quality on fruit color. J Am Soc Hortic Sci 120:785–789
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Fischer TC, Gosch C, Pfeiffer J, Halbwirth H, Halle C, Stich K, Forkmann G (2007) Flavonoid genes of pear (Pyrus communis). Trees 21:521–529
Goff SA, Cone KC, Chandler VL (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6:864–875
Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68:955–964
Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci U S A 97:13579–13584
Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B (1998) Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol 36:741–754
Hironori M, Fumiaki O, Kazuhito S, Hiromi H, Yuzo M (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Jia HJ, Araki A, Okamoto G (2005) Influence of fruit bagging on aroma volatiles and skin coloration of ‘Hakuho’ peach (Prunus persica Batsch). Post Harvest Biol Technol 35:61–68
Jiang CZ, Gu X, Peterson T (2004) Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol 5:R46
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Kano M, Takayanagi T, Harada K, Makino K, Ishikawa F (2005) Antioxidative activity of anthocyanins from purple sweet potato, Ipomoea batatas cultivar Ayamurasaki. Biosci Biotechnol Biochem 69:979–988
Kanokporn S, Yukiko M, Mami Y, Kazuki S (2002) AWD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol Biol 50:485–495
Kataoka I, Beppu K (2004) UV irradiance increases development of red skin color and anthocyanins in ‘Hakuho’ peach. HortScience 39:1234–1237
Kim SH, Lee JR, Hong ST, Yoo YK, An G, Kim SR (2003) Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 165:403–413
Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215:924–933
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Morishima A (1998) Identification of preferred binding sites of a light-inducible DNA-binding factor (MNF1) within 5′-upstream sequence of C4-type phosphoenolpyruvate carboxylase gene in maize. Plant Mol Biol 38:633–646
Pirie A, Mullins MG (1976) Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate abscisic acid. Plant Physiol 58:468–472
Ramsay NA, Walker AR, Mooney M, Gray JC (2003) Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol Biol 152:679–688
Ren QP, Zhang FJ, Lü FT, Wang GY, Zhu FZ, Xing ZD (2007) Breeding of a new red pear-Aoguan. China Fruits 6:12–13
Sainz MB, Grotewold E, Chandler VL (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9:611–625
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18:831–851
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Shozo K, Nami GY, Hirohiko H (2004) Retrotransposon-induced mutations in grape skin color. Science 304:982
Spelt C, Quattrocchio F, Mol JNM, Koes R (2000) Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12:1619–1631
Steyn WJ, Holcroft DM, Wand SJE, Jacobs G (2004a) Regulation of pear color development in relation to activity of flavonoid enzymes. J Am Soc Hortic Sci 129:6–12
Steyn WJ, Holcroft DM, Wand SJE, Jacobs G (2004b) Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J Am Soc Hortic Sci 129:13–19
Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2005) Red colour development and loss in pears. Acta Hortic 671:79–85
Suda I, Oki T, Masuda M, Kobayashi M, Nishiba Y, Furuta S (2003) Physiological functionality of purple-fleshed sweet potatoes containing anthocyanins and their utilization in foods. JARQ 37:167–173
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Tao B, Shu Q, Wang JJ, Zhang WB (2004) Studies on frontiers and prospects of resources of red pears and its application. Southwest Chin J Agric Sci 47:409–412
Tsuda T, Yamaguchi M, Honda C, Moriguchi T (2004) Expression of anthocyanin biosynthesis genes in the skin of peach and nectarine fruit. J Am Soc Hortic Sci 129:857–862
Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139:806–821
Walker AR, Elizabeth L, Jochen B, Debra AJ, Mc D, Mark RT, Simon PR (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49:772–785
Wang YL (1997) Study report of red pear breeding. Fruit Sci 14:71–76
Wang HC, Huang XM, Hu GB, Huang HB (2004) Studies on the relationship between anthocyanin biosynthesis and related enzymes in Litchi pericarp. Sci Agric Sin 37:2028–2032
Winkel SB (2001) Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol 126:485–493
Yao GL, Yuan LC (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 43:649–656
Zhang WB, Zhang JR (1993) Red pears resources in subtropical of Yunnan. China Fruits 4:16–17
Zhang WB, Zhang JR, Li XL, Shu Q, Zeng YW (1997) Germplasm resources of red pears in Yunnan and its application. South China Fruits 26:38–39
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Zou YP, Ge S, Wang XD (eds) (2001) The molecular markers in systematic and evolutionary botany. Science Press, Beijing, pp 16–17
Acknowledgments
We thank Aoguan Fruitco Ltd. for kindly supplying pear fruit, and for harvesting and packaging of fruit. We also thank Dao-Lin Fu, Xian-Sheng Zhang, Xiang-Yu Zhao, Xiang-Bin Chen, Yu-Xiao Su, Shuai-Shuai Wang, Xiao-Liu Chen, Yu-Jin Hao, Shuang-Shuang Wang and Xing-Bin Xie for their help. This study was supported by the National 863 Program of China (2006AA100108).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, S., Wang, Y., Yang, S. et al. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255 (2010). https://doi.org/10.1007/s00425-010-1170-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1170-5