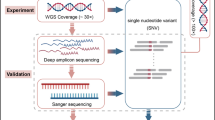
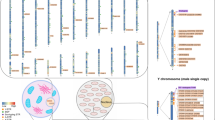
Abstract
Differentiating between monozygotic (MZ) twins remains difficult because they have the same genetic makeup. Applying the traditional STR genotyping approach cannot differentiate one from the other. Heteroplasmy refers to the presence of two or more different mtDNA copies within a single cell and this phenomenon is common in humans. The levels of heteroplasmy cannot change dramatically during transmission in the female germ line but increase or decrease during germ-line transmission and in somatic tissues during life. As massively parallel sequencing (MPS) technology has advanced, it has shown the extraordinary quantity of mtDNA heteroplasmy in humans. In this study, a probe hybridization technique was used to obtain mtDNA and then MPS was performed with an average sequencing depth of above 4000. The results showed us that all ten pairs of MZ twins were clearly differentiated with the minor heteroplasmy threshold at 1.0%, 0.5%, and 0.1%, respectively. Finally, we used a probe that targeted mtDNA to boost sequencing depth without interfering with nuclear DNA and this technique can be used in forensic genetics to differentiate the MZ twins.






Similar content being viewed by others
References
Oosthuizen T, Howes LM (2022) The development of forensic DNA analysis: new debates on the issue of fundamental human rights. Forensic Sci Int Genet 56:102606
Nwawuba Stanley U et al (2020) Forensic DNA profiling: autosomal short tandem repeat as a prominent marker in crime investigation. Malays J Med Sci 27(4):22–35
Xu QN, Li CT, Liu XL (2018) Research progress on discrimination of monozygotic twins. Fa Yi Xue Za Zhi 34(6):672–677
Yang YR et al (2012) Progress on epigenetics applications in forensic science. Fa Yi Xue Za Zhi 28(5):366–370
Vidaki A, Daniel B, Court DS (2013) Forensic DNA methylation profiling--potential opportunities and challenges. Forensic Sci Int Genet 7(5):499–507
Marqueta-Gracia JJ et al (2018) Differentially methylated CpG regions analyzed by PCR-high resolution melting for monozygotic twin pair discrimination. Forensic Sci Int Genet 37:e1–e5
Castellani CA et al (2014) Biological relevance of CNV calling methods using familial relatedness including monozygotic twins. BMC Bioinformatics 15:114
Planterose Jimenez B et al (2021) Equivalent DNA methylation variation between monozygotic co-twins and unrelated individuals reveals universal epigenetic inter-individual dissimilarity. Genome Biol 22(1):18
Xiao C et al (2019) Differences of microRNA expression profiles between monozygotic twins’ blood samples. Forensic Sci Int Genet 41:152–158
Nistico L et al (2006) Concordance, disease progression, and heritability of coeliac disease in Italian twins. Gut 55(6):803–808
Sites ER et al (2017) Analysis of copy number variants in 11 pairs of monozygotic twins with neurofibromatosis type 1. Am J Med Genet A 173(3):647–653
Stewart JB, Chinnery PF (2021) Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat Rev Genet 22(2):106–118
Anderson S et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290(5806):457–465
Chiaratti MR, Chinnery PF (2022) Modulating mitochondrial DNA mutations: factors shaping heteroplasmy in the germ line and somatic cells. Pharmacol Res 185:106466
Uchiumi T, Kang D (2012) The role of TFAM-associated proteins in mitochondrial RNA metabolism. Biochim Biophys Acta 1820(5):565–570
Yasukawa T, Kang D (2018) An overview of mammalian mitochondrial DNA replication mechanisms. J Biochem 164(3):183–193
Kennedy SR et al (2013) Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet 9(9):e1003794
Legati A et al (2021) Current and new next-generation sequencing approaches to study mitochondrial DNA. J Mol Diagn 23(6):732–741
Mertens J et al (2019) Detection of heteroplasmic variants in the mitochondrial genome through massive parallel sequencing. Bio Protoc 9(13):e3283
Wang Z et al (2015) Differentiating between monozygotic twins through next-generation mitochondrial genome sequencing. Anal Biochem 490:1–6
McElhoe JA et al (2014) Development and assessment of an optimized next-generation DNA sequencing approach for the mtgenome using the Illumina MiSeq. Forensic Sci Int Genet 13:20–29
Yu H et al (2022) Hot spots-making directed evolution easier. Biotechnol Adv 56:107926
Chen L et al (2020) Highly accurate mtGenome haplotypes from long-read SMRT sequencing can distinguish between monozygotic twins. Forensic Sci Int Genet 47:102306
Gaudin M, Desnues C (2018) Hybrid capture-based next generation sequencing and its application to human infectious diseases. Front Microbiol 9:2924
Shih, S.Y., et al., Correction: Shelly Y. Shih; et al.; Applications of probe capture enrichment next generation sequencing for whole mitochondrial genome and 426 nuclear SNPs for forensically challenging samples. Genes 2018, 9, 49.
Yin L et al (2021) Validation of the Microreader 28A ID System: A 6-dye multiplex amplification assay for forensic application. Electrophoresis 42(19):1928–1935
Shih SY et al (2018) Applications of probe capture enrichment next generation sequencing for whole mitochondrial genome and 426 nuclear SNPs for forensically challenging samples. Genes (Basel) 9(1)
Lang J et al (2021) Evaluation of the MGISEQ-2000 sequencing platform for illumina target capture sequencing libraries. Front Genet 12:730519
Chen S et al (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
McKenna A et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Li H et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079
Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10(10):1556–1566
Bensasson D et al (2001) Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol 16(6):314–321
Parakatselaki ME, Ladoukakis ED (2021) mtDNA heteroplasmy: origin, detection, significance, and evolutionary consequences. Life (Basel) 11(7):633
Vidaki A et al (2017) Epigenetic discrimination of identical twins from blood under the forensic scenario. Forensic Sci Int Genet 31:67–80
Xu J et al (2015) LINE-1 DNA methylation: a potential forensic marker for discriminating monozygotic twins. Forensic Sci Int Genet 19:136–145
Zhang N et al (2015) Intra-monozygotic twin pair discordance and longitudinal variation of whole-genome scale DNA methylation in adults. PLoS One 10(8):e0135022
Vidaki A et al (2018) Investigating the epigenetic discrimination of identical twins using buccal swabs, saliva, and cigarette butts in the forensic setting. Genes (Basel) 9(5):252
Fang C et al (2019) MicroRNA profile analysis for discrimination of monozygotic twins using massively parallel sequencing and real-time PCR. Forensic Sci Int Genet 38:23–31
Li D et al (2022) Advances in bioactivity of MicroRNAs of plant-derived exosome-like nanoparticles and milk-derived extracellular vesicles. J Agric Food Chem 70(21):6285–6299
Liang H et al (2014) The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip Rev RNA 5(2):285–300
Rolf B, Krawczak M (2021) The germlines of male monozygotic (MZ) twins: Very similar, but not identical. Forensic Sci Int Genet 50:102408
Burr SP, Chinnery PF (2022) Measuring single-cell mitochondrial DNA copy number and heteroplasmy using digital droplet polymerase chain reaction. J Vis Exp 185:e63870
Abd Radzak SM et al (2022) Insights regarding mitochondrial DNA copy number alterations in human cancer (Review). Int J Mol Med 50(2):1–8
Wang Y et al (2022) Genetic landscape of human mitochondrial genome using whole-genome sequencing. Hum Mol Genet 31(11):1747–1761
Dulovic-Mahlow M et al (2021) Discordant monozygotic parkinson disease twins: role of mitochondrial integrity. Ann Neurol 89(1):158–164
Yin D et al (2022) Gapless genome assembly of East Asian finless porpoise. Sci Data 9(1):765
Sun Y et al (2021) Characterizing sensitivity and coverage of clinical WGS as a diagnostic test for genetic disorders. BMC Med Genomics 14(1):102
Lee J, Jung J (2022) First record of the complete mitochondrial genome of Tubifex tubifex (Muller) 1774 (Annelida; Clitellata; Oligochaeta) and phylogenetic analysis. Mitochondrial DNA B Resour 7(7):1208–1210
Jeon SA et al (2021) Comparison between MGI and Illumina sequencing platforms for whole genome sequencing. Genes Genomics 43(7):713–724
Zhu K et al (2021) Comparative Performance of the MGISEQ-2000 and Illumina X-Ten Sequencing Platforms for Paleogenomics. Front Genet 12:745508
Korostin D et al (2020) Comparative analysis of novel MGISEQ-2000 sequencing platform vs Illumina HiSeq 2500 for whole-genome sequencing. PLoS One 15(3):e0230301
Bardan F, Higgins D, Austin JJ (2023) A custom hybridisation enrichment forensic intelligence panel to infer biogeographic ancestry, hair and eye colour, and Y chromosome lineage. Forensic Sci Int Genet 63:102822
Bose N et al (2018) Target capture enrichment of nuclear SNP markers for massively parallel sequencing of degraded and mixed samples. Forensic Sci Int Genet 34:186–196
Tu J et al (2015) Systematic characteristic exploration of the chimeras generated in multiple displacement amplification through next generation sequencing data reanalysis. PLoS One 10(10):e0139857
Data availability
The data sets used and/or analyzed during this study are available from the corresponding author upon reasonable request.
Funding
This study was supported by the Natural Science Foundation of Liaoning Province (2022-YGJC-45) and the Student Innovation and Entrepreneurship Training Program of China Medical University in 2023.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of China Medical University (Shenyang, Liaoning Province, People’s Republic of China).
Disclosure of potential conflicts of interest
The authors declare that they have no competing interest.
Informed consent
All the participants were included after providing written informed consent.
Research involving human participants and/or animals
This study involves human participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Zhong, Kuo Zeng, and Atif Adnan are the co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, Y., Zeng, K., Adnan, A. et al. Discrimination of monozygotic twins using mtDNA heteroplasmy through probe capture enrichment and massively parallel sequencing. Int J Legal Med 137, 1337–1345 (2023). https://doi.org/10.1007/s00414-023-03033-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-023-03033-x