Abstract
Purpose
Tuberous sclerosis complex (TSC) is an autosomal dominant multisystem disorder characterized by hamartomas in multiple organ systems. The TSC1 and TSC2 genes have been identified as the genetic basis of TSC. Two gene tests were used for definitive genetic diagnosis.
Methods
In our study, the case of a Chinese pediatric patient with seizures, hypomelanotic macules, hyperpigmented patches, multiple parenchymal lesions in the ventricle, and developmental retardation is detailed. Whole-genome sequencing (WGS) and multiplex ligation-dependent probe amplification (MLPA) were employed to detect genetic variations and copy number variations of TSC1 and TSC2.
Results
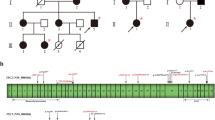
A novel heterozygous nonsense mutation in the TSC2 gene (c.3751A>T, p.Lys1251Ter) was identified in a Chinese pediatric patient suffering from TSC, whose unaffected parents did not carry this mutation. The mutation was classified as “pathogenic” according to the American College of Medical Genetics (ACMG) guidelines.
Conclusion
WGS was carried out to definitively diagnose and detect variations in the exon and noncoding region of the gene and copy number variations in the whole genome simultaneously. For diseases with complex genetic mechanisms, WGS as the first-line test can be efficient and cost-effective for clinical diagnosis.


Similar content being viewed by others
References
Krueger DA, Northrup H, Roberds S, Smith K, Sampson J, Korf B, Kwiatkowski DJ, Mowat D, Nellist M, Povey S (2013) Tuberous sclerosis complex surveillance and management: recommendations of the 2012 International Tuberous Sclerosis Complex Consensus Conference. Pediatr Neurol 49:255–265. https://doi.org/10.1016/j.pediatrneurol.2013.08.001
Rosset C, Netto CBO, Ashton-Prolla P (2017) TSC1 and TSC2 gene mutations and their implications for treatment in tuberous sclerosis complex: a review. Genet Mol Biol 40:69–79. https://doi.org/10.1590/1678-4685-gmb-2015-0321
Hung C-C, Su Y-N, Chien S-C, Liou H-H, Chen C-C, Chen P-C, Hsieh C-J, Chen C-P, Lee W-T, Lin W-L (2006) Molecular and clinical analyses of 84 patients with tuberous sclerosis complex. BMC Med Genet 7:72. https://doi.org/10.1186/1471-2350-7-72
Sasongko TH, Ismail NFD, Malik NMANA, Zabidi-Hussin Z (2015) Rapamycin and its analogues (rapalogs) for tuberous sclerosis complex-associated tumors: a systematic review on non-randomized studies using meta-analysis. Orphanet J Rare Dis 10:95. https://doi.org/10.1186/s13023-015-0317-7
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD (2018) ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am J Hum Genet 103:474–483. https://doi.org/10.1016/j.ajhg.2018.08.005
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423. https://doi.org/10.1038/gim.2015.30
Tyburczy ME, Dies KA, Glass J, Camposano S, Chekaluk Y, Thorner AR, Lin L, Krueger D, Franz DN, Thiele EA (2015) Mosaic and intronic mutations in TSC1/TSC2 explain the majority of TSC patients with no mutation identified by conventional testing. PLoS Genet 11:e1005637. https://doi.org/10.1371/journal.pgen.1005637
Nellist M, Brouwer RW, Kockx CE, van Veghel-Plandsoen M, Withagen-Hermans C, Prins-Bakker L, Hoogeveen-Westerveld M, Mrsic A, van den Berg MM, Koopmans AE (2015) Targeted next generation sequencing reveals previously unidentified TSC1 and TSC2 mutations. BMC Med Genet 16:10. https://doi.org/10.1186/s12881-015-0155-4
Crino PB, Nathanson KL, Henske EP (2006) The tuberous sclerosis complex. N Engl J Med 355:1345–1356. https://doi.org/10.1056/nejmra055323
Sancak O, Nellist M, Goedbloed M, Elfferich P, Wouters C, Maat-Kievit A, Zonnenberg B, Verhoef S, Halley D, van den Ouweland A (2005) Mutational analysis of the TSC1 and TSC2 genes in a diagnostic setting: genotype–phenotype correlations and comparison of diagnostic DNA techniques in tuberous sclerosis complex. Eur J Hum Genet 13:731–741. https://doi.org/10.1038/sj.ejhg.5201402
Mayer K, Fonatsch C, Wimmer K, Van Den Ouweland AM, Maat-Kievit AJ (2014) Clinical utility gene card for: tuberous sclerosis complex (TSC1, TSC2). Eur J Hum Genet 22:293. https://doi.org/10.1038/ejhg.2013.129
Dabora SL, Jozwiak S, Franz DN, Roberts PS, Nieto A, Chung J, Choy Y-S, Reeve MP, Thiele E, Egelhoff JC (2001) Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am J Hum Genet 68:64–80. https://doi.org/10.1086/316951
Strizheva GD, Carsillo T, Kruger WD, Sullivan EJ, Ryu JH, Henske EP (2001) The spectrum of mutations in TSC1 and TSC2 in women with tuberous sclerosis and lymphangiomyomatosis. Am J Respir Crit Care Med 163:253–258. https://doi.org/10.1164/ajrccm.163.1.2005004
Ismail NFD, Rani AQ, Malik NMANA, Hock CB, Azlan SNM, Razak SA, Keng WT, Ngu LH, Silawati AR, Yahya NA (2017) Combination of multiple ligation-dependent probe amplification and illumina MiSeq amplicon sequencing for TSC1/TSC2 gene analyses in patients with tuberous sclerosis complex. J Mol Diagn 19:265–276. https://doi.org/10.1016/j.jmoldx.2016.10.009
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81971219).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was approved by the Ethics Committee of Xinqiao Hospital. Informed consent to this study was obtained from the parents of the patient.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, Mh., Wang, Zk., Huang, Y. et al. A novel de novo TSC2 nonsense mutation detected in a pediatric patient with tuberous sclerosis complex. Childs Nerv Syst 37, 253–257 (2021). https://doi.org/10.1007/s00381-020-04702-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00381-020-04702-7