Abstract
Plants in the family Solanaceae are used as model systems in comparative and evolutionary genomics. The complete chloroplast genomes of seven solanaceous species have been sequenced, including tobacco, potato and tomato, but not peppers. We analyzed the complete chloroplast genome sequence of the hot pepper, Capsicum annuum. The pepper chloroplast genome was 156,781 bp in length, including a pair of inverted repeats (IR) of 25,783 bp. The content and the order of 133 genes in the pepper chloroplast genome were identical to those of other solanaceous plastomes. To characterize pepper plastome sequence, we performed comparative analysis using complete plastome sequences of pepper and seven solanaceous plastomes. Frequency and contents of large indels and tandem repeat sequences and distribution pattern of genome-wide sequence variations were investigated. In addition, a phylogenetic analysis using concatenated alignments of coding sequences was performed to determine evolutionary position of pepper in Solanaceae. Our results revealed two distinct features of pepper plastome compared to other solanaceous plastomes. Firstly, large indels, including insertions on accD and rpl20 gene sequences, were predominantly detected in the pepper plastome compared to other solanaceous plastomes. Secondly, tandem repeat sequences were particularly frequent in the pepper plastome. Taken together, our study represents unique features of evolution of pepper plastome among solanaceous plastomes.







Similar content being viewed by others
References
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bohs L, Olmstead RG (1997) Phylogenetic relationships in Solanum (Solanaceae) based on ndhF sequences. Syst Bot 22:5–17
Bortiri E, Coleman-Derr D, Lazo GR, Anderson OD, Gu YQ (2008) The complete chloroplast genome sequence of Brachypodium distachyon: sequence comparison and phylogenetic analysis of eight grass plastomes. BMC Res Notes 1:61
Calsa Junior T, Carraro DM, Benatti MR, Barbosa AC, Kitajima JP, Carrer H (2004) Structural features and transcript-editing analysis of sugarcane (Saccharum officinarum L.) chloroplast genome. Curr Genet 46:366–373
Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK (2006) The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol 23:2175–2190
Chung HJ, Jung JD, Park HW, Kim JH, Cha HW, Min SR, Jeong WJ, Liu JR (2006) The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep 25:1369–1379
Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, Meyer L, Wilson RK, Newton KJ (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136:3486–3503
Cosner ME, Jansen RK, Palmer JD, Downie SR (1997) The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet 31:419–429
Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK (2006) Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet 112:1503–1518
Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ (1992) Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc Natl Acad Sci USA 89:7722–7726
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279
Gao L, Yi X, Yang YX, Su YJ, Wang T (2009) Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol 9:130
Gillman JD, Bentolila S, Hanson MR (2007) The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J 49:217–227
Goffinet B, Wickett NJ, Werner O, Ros RM, Shaw AJ, Cox CJ (2007) Distribution and phylogenetic significance of the 71-kb inversion in the plastid genome in Funariidae (Bryophyta). Ann Bot 99:747–753
Gray MW (1989) The evolutionary origins of organelles. Trends Genet 5:294–299
Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, Haberer G, Herrmann RG (2008) The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. sequence evaluation and plastome evolution. Nucleic Acids Res 36:2366–2378
Guo X, Castillo-Ramirez S, Gonzalez V, Bustos P, Fernandez-Vazquez JL, Santamaria RI, Arellano J, Cevallos MA, Davila G (2007) Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8:228
Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66:350–361
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui L (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol 395:348–384
Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H (2006) Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6:32
Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104:19369–19374
Jo YD, Jeong HJ, Kang BC (2009) Development of a CMS-specific marker based on chloroplast-derived mitochondrial sequence in pepper. Plant Biotechnol Rep 3:309–315
Kahlau S, Aspinall S, Gray JC, Bock R (2006) Sequence of the tomato chloroplast DNA and evolutionary comparison of Solanaceous plastid genomes. J Mol Evol 63:194–207
Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S (2000) Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res 7:323–330
Khakhlova O, Bock R (2006) Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J 46:85–94
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean Ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Kim KJ, Lee HL (2005) Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells 19:104–113
Kim DH, Kang JG, Kim BD (2007) Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol Biol 63:519–532
Kim HJ, Baek KH, Lee SW, Kim J, Lee BW, Cho HS, Kim WT, Choi D, Hur CG (2008) Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol 8:101
Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44:237–244
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105:2923–2928
Lee SS, Jeong WJ, Bae JM, Bang JW, Liu JR, Harn CH (2004) Characterization of the plastid-encoded carboxyltransferase subunit (accD) gene of potato. Mol Cells 17:422–429
Leseberg CH, Duvall MR (2009) The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol 69:311–318
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Maier RM, Neckermann K, Igloi GL, Kossel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251:614–628
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Melotto-Passarin DM, Berger IJ, Dressano K, De Martin VF, Oliveira GCX, Bock R, Carrer H (2008) Phylogenetic relationships in Solanaceae and related species based on cpDNA sequence from plastid trnE–trnT region. Crop Breed Appl Biotechnol 8:85–95
Michelangeli FA, Davis JI, Stevenson DW (2003) Phylogenetic relationships among Poaceae and related families as inferred from morphology, inversions in the plastid genome, and sequence data from the mitochondrial and plastid genomes. Am J Bot 90:93–106
Milligan BG, Hampton JN, Palmer JD (1989) Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol 6:355–368
Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA 107:4623–4628
Ogihara Y, Terachi T, Sasakuma T (1988) Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci USA 85:8573–8577
Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N et al (2002) Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol Genet Genomics 266:740–746
Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM (2008) A molecular phylogeny of the Solanaceae. Taxon 57:1159–1181
Palmer JD (1985) Comparative organization of chloroplast genomes. Annu Rev Genet 19:325–354
Palmer JD (1990) Contrasting modes and tempos of genome evolution in land plant organelles. Trends Genet 6:115–120
Palmer JD, Nugent JM, Herbon LA (1987) Unusual structure of geranium chloroplast DNA: a triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc Natl Acad Sci USA 84:769–773
Park J, Park B, Jung K, Jang S, Yu K, Choi J, Kong S, Park J, Kim S, Kim H, Kim S, Kim JF, Blair JE, Lee K, Kang S, Lee YH (2008) CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res 36:D562–D571
Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK (2005) Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol 59:309–322
Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM (2002) The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol 19:1602–1612
Shimada H, Sugiura M (1989) Pseudogenes and short repeated sequences in the rice chloroplast genome. Curr Genet 16:293–301
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5:2043–2049
Sugiura M (1989) The chloroplast chromosomes in land plants. Annu Rev Cell Biol 5:51–70
Sugiura M, Hirose T, Sugita M (1998) Evolution and mechanism of translation in chloroplasts. Annu Rev Genet 32:437–459
Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272:603–615
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. In: Current protocols in bioinformatics, chap 2, unit 2.3
Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci USA 91:9794–9798
Wang Y, Diehl A, Wu F, Vrebalov J, Giovannoni J, Siepel A, Tanksley SD (2008) Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180:391–408
Wilson AC, Ochman H, Prager EM (1987) Molecular time scale for evolution. Trends Genet 3:241–247
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Yukawa M, Tsudzuki T, Sugiura M (2006) The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum. Mol Genet Genomics 275:367–373
Acknowledgments
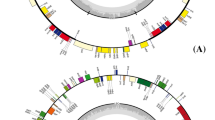
This work was supported by a grant (code20070301034022) from the BioGreen21 program, by the Rural Development Administration and by the ARC program supported by Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, the Republic of Korea. We thank to Kim, Suk-Jun who kindly helped us with drawing a gene map of pepper chloroplast genome.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Rights and permissions
About this article
Cite this article
Jo, Y.D., Park, J., Kim, J. et al. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep 30, 217–229 (2011). https://doi.org/10.1007/s00299-010-0929-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0929-2