Abstract
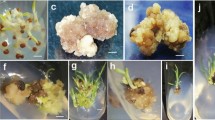
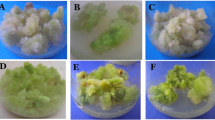
Two protocols of plant regeneration for cotton were adopted in this study, namely, 2, 4-D and kinetin hormone combination and IBA and kinetin hormone combination. Twenty-eight embryogenic cell lines via somatic embryogenesis and 67 regenerated plants from these embryogenic calli were selected and used for random amplified polymorphic DNA (RAPD), simple sequence repeat (SSR), chromosomal number counting, and flow cytometric analysis. The roles of RAPD and SSR markers in detecting somaclonal variation of cotton (Gossypium hirsutum L.) were evaluated. Two cluster analyses were performed to express, in the form of dendrograms, the relationships among the hormone combinations and the genetic variability. Both DNA-based techniques were able to amplify all of the cell clones and regenerated plantlets genomes and relative higher genetic variation could be detected in the culture type with 2, 4-D and kinetin hormone combination. The result suggested that 2, 4-D and kinetin hormone combination could induce relative high somaclonal variation and RAPD and SSR markers are useful in detecting somaclonal variation of regenerated cotton plants via somatic embryogenesis. Chromosome number counting and flow cytometry analysis revealed that the number of chromosomes and ploidy levels were nearly stable in all regenerated plants except two regenerated plantlets (lost 4 and 5 chromosomes, respectively) which meant that cytological changes were not correlated with the frequency of RAPD and SSR polymorphisms. This result also might mean that the cell lines with variation of chromosome numbers were difficult to regenerate plants.











Similar content being viewed by others
References
Bouman H, Deklerk GJ (2001) Measurement of the extent of somaclonal variation in Begonia plants regenerated under various conditions: comparison of three assays. Theor Appl Genet 102:111–117
Conde P, Loureiro J, Santos C (2004) Somatic embryogenesis and plant regeneration from leaves of Ulmus minor Mill. Plant Cell Rep 22:632–639
Davidonis GH, Hamilton RH (1983) Plant regeneration from callus tissue of Gossypium hirsutum L. Plant Sci Lett 32:89–93
DeVerno LL, Charest PJ, Bonen L (1994) Mitochondrial DNA variation in somatic embryogenic cultures of Larix. Theor Appl Genet 88:727–732
Dolezel J (1991) Flow cytometric analysis of nuclear DNA content in higher plants. Phytochem Anal 2:143–154
Dolezel J (1997) Applications of flow cytometry for the study of plant genomes. J Appl Genet 38:285–302
Dolezel J, Lucretti S, Novak FJ (1987) The influence of 2, 4-Dichlorophenoxyacetic acid on cell cycle kinetics and sisterchromatid exchange frequency in garlic (Allium sativum) meristem cells. Biol Plant (Prague) 29:253–257
Dolezel J, Bartos J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry Part A 51A:127–128
El Maataoui M, Pichot C (1999) Nuclear and cell fusion cause polyploidy in the mega gametophyte of common cypress, Cupressus sempervirens L. Planta 208:345–351
Firoozabady E, DeBoer DL (1993) Plant regeneration via somatic embryogenesis in many cultivars of cotton (Gossypim hirsutum L.). In Vitro Cell Dev Biol Plant 29:166–173
Fourré JL, Berger P, Niquet L, Andre P (1997) Somatic embryogenesis and somaclonal variation in Norway spruce: morphogenetic, cytogenetic and molecular approaches. Theor Appl Genet 94:159–169
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension culture of soybean roots cells. Exp Cell Res 50:150–158
He DH, Lin ZX, Zhang XL, Nie YC, Guo XP, Zhang YX (2007) QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of G. hirsutum × G. barbadense. Euphytica 153:181–197
Hendrix B, Steward JM (2005) Estimation of the nuclear DNA content of Gossypium species. Ann Bot 95:789–797
Isabel N, Tremblay L, Michaud M, Tremblay FM, Bousquet J (1993) RAPDs as an aid to evaluate the genetic integrity of somatic embryogenesis derived population of Picea mariana (Mill.) B·S.P. Theor Appl Genet 86:81–87
Jin SX, Zhang XL, Liang SG, Nie YC, Guo XP, Huang C (2005) Factors affecting transformation efficiency of embryogenic callus of Upland cotton (Gossypium hirsutum L.) with Agrobacterium tumefaciens. Plant Cell Tiss Org Cult 81:229–237
Jin SX, Zhang XL, Liang SG, Nie YC, Guo XP (2006) An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum L.). Plant Cell Tiss Org Cult 85:181–185
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–88
Ladyzynski M, Burza W, Malepszy S (2002) Relationship between somaclonal variation and type of culture in cucumber. Euphytica 125:349–356
Larkin PJ, Scowcroft WR (1981) Somaclonal variation: a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:197–214
Lin Z, Zhang X, Nie Y, Guo X, Feng C, JMcD Stewart (2005) A genetic linkage map of tetraploid cotton constructed by SSR, SRAP and RAPD and QTLs mapping for cotton fiber traits. Plant Breed 124:180–187
May RA, Sink KC (1995) Genotype and auxin influence direct somatic embryogenesis from protoplasts derived from embryogenic cell suspensions of Asparagus officinalis L. Plant Sci 108:71–84
Moscone EA, Baranyi M, Ebert I, Greilhuber J, Ehrendorfer F, Hunziker AT (2003) Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Ann Bot (Lond) 92:21–29
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissues cultures. Physiol Planta 15:473–479
Murata M (1989) Effects of auxin and cytokinin on induction of sister chromatid exchanges in cultured cells of wheat (Triticum aestivum L.). Theor Appl Genet 78:521–524
Ngezahayo F, Dong YS, Liu B (2007) Somaclonal variation at the nucleotide sequence level in rice (Oryza sativa L.) as revealed by RAPD and ISSR markers, and by pairwise sequence analysis. J Appl Genet 48:329–336
Osipova ES, Koveza OV, Troitskiĭ AV, IuI Dolgikh, Shamina ZB, Gostimskiĭ SA (2003) Analysis of specific RAPD- and ISSR-fragments in somaclonal maize (Zea mays L.) and development of SCAR markers based on them. Genetika 39:1664–1672
Palombi MA, Damiano C (2002) Comparison between RAPD and SSR molecular markers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev). Plant Cell Rep 20:1061–1066
Pinto G, Loureiro J, Lopes T, Santos C (2004) Analysis of the genetic stability of Eucalyptus globulus Labill somatic embryos by flow cytometry. Theor Appl Genet 109:580–587
Rahman MH, Rajora OP (2001) Microsatellite DNA somaclonal variation in micropropagated trembling aspen (Populus tremuloides). Plant Cell Rep 20:531–536
Raimondi JP, Masuelli RW, Camadro EL (2001) Assessment of somaclonal variation in Asparagus by RAPD fingerprinting and cytogenetic analyses. Sci Horti 90:19–29
Reuther G, Becker U (1987) Relationships between ploidy variations and organgenic potency in long-term callus cultures of (Asparagus officinalis). Acta Horti 212:143–149
Rival A, Bertrand L, Beule T, Combes MC, Troutslot P, Lashermes P (1998) Suitability of RAPD analysis for the detection of somaclonal variants in oil palm (Elaeiguineensis Jacq). Plant Breed 117:73–76
Rohlf FJ (1993) NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Publishing Ltd, New York
Rohlf FJ (1997) NTSYSpc: numerical taxonomy and multivariate analysis system, version 2.02. Exeter Software, Setauket, New York
Shoemaker RC, Couche LJ, Galbraith DW (1986) Characterization of somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L.). Plant Cell Rep 5:178–181
Stelly DM, Altman DW, Kohel RJ, Rangan TS, Commiskey E (1989) Cytogenetic abnormalities of cotton somaclonal from callus cultures. Genome 32:762–770
Trehin C, Planchais S, Glab N, Perennes C, Tregear J, Bergounioux C (1998) Cell cycle regulation by plant growth regulators: involvement of auxin and cytokinin in the re-entry of Petunia protoplasts into the cell cycle. Planta 206:215–224
Trolinder NL, Chen ZX (1989) Genotype specificity of the somatic embryogenesis response in cotton. Plant Cell Rep 8:133–136
Trolinder NL, Goodin JR (1987) Somatic embryogenesis and plant regeneration in Gossypium hirsutum L. Plant Cell Rep 6:231–234
Trolinder NL, Goodin JR (1988) Somatic embryogenesis in cotton (Gossypium) I. Effects of explants and hormone regime. Plant Cell Tiss Org Cult 12:31–42
Umbeck F, Steward JM (1985) Substitution of cotton cytoplasm from wild diploid species for cotton germplasm improvement. Crop Sci 25:1015–1019
Voo KS, Rugh CL, Kamalay JC (1991) Indirect somatic embryogenesis and plant recovery from cotton. In vitro Cell Dev Bio Plant 27:117–124
Wilkins T, Rajasekaran K, Anderson DM (2000) Cotton biotechnology. Crit Rev Plant Sci 19:511–550
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Wu JH, Zhang XL, Nie YC, Jin SX, Liang SG (2004) Factors affecting somatic embryogenesis and plant regeneration from a range of recalcitrant genotypes of Chinese cottons (Gossypium hirsutum L.). In vitro Cell Dev Bio Plant 40:371–375
Yang H, Tabei Y, Kamada H, Kayano T, Takaiwa F (1999) Detection of somaclonal variation in cultured rice cells using digoxigenin-based random amplified polymorphic DNA. Plant Cell Rep 18:520–526
Zhang XL, Sun JZ, Liu JL (1991) Somatic embryogenesis and plant regeneration in upland cotton. Acta Genetica Sinica 18:461–467
Zhang XL, Zhang JM, Yiao MJ, Sun JZ, Liu JL (1993) Studies on somatic cell culture and its application in cotton (Gossypium hirsutum L.) improvement. J Huazhong Agri Univ 12:421–426
Acknowledgments
The funds supported by National Natural Science Foundation of China (No. 30771368), National High-Tech program (20060AA00105) and China Postdoctoral Science Foundation (20070410949) were appreciated. We also thank Prof. V. Ravishankar Rai, Department of Microbiology, University of Mysore, India for critically evaluating this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Lu.
Rights and permissions
About this article
Cite this article
Jin, S., Mushke, R., Zhu, H. et al. Detection of somaclonal variation of cotton (Gossypium hirsutum) using cytogenetics, flow cytometry and molecular markers. Plant Cell Rep 27, 1303–1316 (2008). https://doi.org/10.1007/s00299-008-0557-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0557-2