Abstract
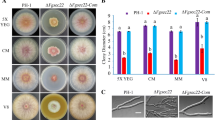
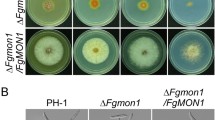
Ubiquitinated biosynthetic and surface proteins destined for degradation are sorted into the lysosome/vacuole via the multivesicular body sorting pathway, which depends on the function of ESCRT machinery. Fusarium head blight (FHB) caused by Fusarium graminearum is one of the most devastating diseases for wheat and barley worldwide. To better understand the role of ESCRT machinery in F. graminearum, we investigated the function of ESCRT-III accessory proteins FgVps60, FgDid2 and FgIst1 in this study. FgVps60–GFP, FgDid2–GFP and FgIst1–GFP are localized to punctate structures adjacent to the vacuolar membrane except for FgIst1–GFP that is also found in the nucleus. Then, the gene deletion mutants ΔFgvps60, ΔFgdid2 and ΔFgist1 displayed defective growth to a different extent. ΔFgvps60 and ΔFgdid2 but not ΔFgist1 also showed significant reduction in hydrophobicity on cell surface, conidiation, perithecia production and virulence. Interestingly, ΔFgist1 mutant produced a significantly higher level of DON while showing a minor reduction in pathogenicity. Microscopic analyses revealed that FgVps60 but not FgIst1 and FgDid2 is necessary for endocytosis. Moreover, spontaneous mutations were identified in the ΔFgvps60 mutant that partially rescued its defects in growth and conidiation. Taken together, we conclude that ESCRT-III accessory proteins play critical roles in growth, reproduction and plant infection in F. graminearum.








Similar content being viewed by others
References
Adell MAY, Teis D (2011) Assembly and disassembly of the ESCRT-III membrane scission complex. Febs Lett 585:3191–3196. https://doi.org/10.1016/j.febslet.2011.09.001
Alexander NJ, Proctor RH, Mccormick SP, Abbas HK (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev 28:198–215. https://doi.org/10.1080/15569540903092142
Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ (2008) ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell 14:50–61. https://doi.org/10.1016/j.devcel.2007.10.021
Babst M, Sato TK, Banta LM, Emr SD (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. Embo J 16:1820–1831. https://doi.org/10.1093/emboj/16.8.1820
Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD (2002a) Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3:271–282. https://doi.org/10.1016/S1534-5807(02)00220-4
Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD (2002b) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3:283–289. https://doi.org/10.1016/S1534-5807(02)00219-8
Bai G, Shaner G (2004) Management and resistance in wheat and barley to fusarium head blight. Annu Rev Phytopathol 42:135–161. https://doi.org/10.1146/annurev.phyto.42.040803.140340
Bai GH, Desjardins AE, Plattner RD (2002) Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153:91–98. https://doi.org/10.1023/A:1014419323550
Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol 4:534–539. https://doi.org/10.1038/ncb815
Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC (2003) Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol 163:237–243. https://doi.org/10.1083/jcb.200305007
Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH (2004) Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194–210. https://doi.org/10.1111/j.1600-0854.2004.00169.x
Brown DW, Dyer RB, Mccormick SP, Kendra DF, Plattner RD (2004) Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet Biol 41:454–462. https://doi.org/10.1016/j.fgb.2003.12.002
Cappellini RA, Peterson JL (1965) Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57:962–966. https://www.jstor.org/stable/3756895
Catlett NL, Lee BN, Yoder OC, Turgeon BG (2003) Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50:9–11. https://doi.org/10.4148/1941-4765.1150
Chen J, Zheng W, Zheng S, Zhang D, Sang W, Chen X, Li G, Lu G, Wang Z (2008) Rac1 Is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. Plos Pathog 4:e1000202. https://doi.org/10.1371/journal.ppat.1000202
Chen A, Xie Q, Lin Y, Xu H, Shang W, Zhang J, Zhang D, Zheng W, Li G, Wang Z (2016) Septins are involved in nuclear division, morphogenesis and pathogenicity in Fusarium graminearum. Fungal Genet Biol Fg B 94:79–87. https://doi.org/10.1016/j.fgb.2016.07.005
Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422:37–44. https://doi.org/10.1038/nature01451
De Walle JV, Sergent T, Piront N, Toussaint O, Schneider YJ, Larondelle Y (2010) Deoxynivalenol affects in vitro intestinal epithelial cell barrier integrity through inhibition of protein synthesis. Toxicol Appl Pharmacol 245:291–298. https://doi.org/10.1016/j.taap.2010.03.012
Desiardins AE, Proctor RH, Bai GH, Mccormick SP, Shaner G, Buechley G, Hohn TM (1996) Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol Plant Microbe Interact 9:775–781 Publication no. M-1996-1023-03R
Desjardins AE (2003) Gibberella From A (Venaceae) to Z (EAE). Annu Rev Phytopathol 41:177–198. https://doi.org/10.1146/annurev.phyto.41.011703.115501
Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M (2008) Ist1 regulates Vps4 localization and assembly. Mol Biol Cell 19:465–474. https://doi.org/10.1091/mbc.e07-08-0747
Ding M, Li J, Fan X, He F, Yu X, Chen L, Zou S, Liang Y, Yu J (2018) Aquaporin1 regulates development, secondary metabolism and stress responses in Fusarium graminearum. Curr Genet 64:1057–1069. https://doi.org/10.1007/s00294-018-0818-8
Dou X, Wang Q, Qi Z, Song W, Wang W, Guo M, Zhang H, Zhang Z, Wang P, Zheng X (2011) MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae. Plos One 6:e16439. https://doi.org/10.1371/journal.pone.0016439
Fuchs U, Hause G, Schuchardt I, Steinberg G (2006) Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell 18:2066–2081. https://doi.org/10.1105/tpc.105.039388
Gale LR, Chen LF, Hernick CA, Takamura K, Kistler HC (2002) Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 92:1315–1322. https://doi.org/10.1094/PHYTO.2002.92.12.1315
Gardiner DM, Kazan K, Manners JM (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet Biol 46:604–613. https://doi.org/10.1016/j.fgb.2009.04.004
Goswami RS, Kistler HC (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525. https://doi.org/10.1111/j.1364-3703.2004.00252.x
Gruenberg J, Stenmark H (2004) The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5:317–323. https://doi.org/10.1038/nrm1360
Haag C, Pohlmann T, Feldbrügge M (2017) The ESCRT regulator Did2 maintains the balance between long-distance endosomal transport and endocytic trafficking. Plos Genet 13:e1006734. https://doi.org/10.1371/journal.pgen.1006734
Hanson PI, Cashikar A (2012) Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28:337–362. https://doi.org/10.1146/annurev-cellbio-092910-154152
Henne WM, Stenmark H, Emr SD (2013) Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol 5:1288–1302. https://doi.org/10.1101/cshperspect.a016766
Hou Z, Xue C, Peng Y, Katan T, Kistler HC, Xu JR (2002) A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum Is required for female fertility, heterokaryon formation, and plant infection. Mol Plant-Microbe Interact 15:1119–1127. https://doi.org/10.1094/MPMI.2002.15.11.1119
Islam KT, Bond JP, Fakhoury AM (2017) FvSNF1, the sucrose non-fermenting protein kinase gene of Fusarium virguliforme, is required for cell-wall-degrading enzymes expression and sudden death syndrome development in soybean. Curr Genet 63:723–738. https://doi.org/10.1007/s00294-017-0676-9
Jia LJ, Tang WH (2015) The omics era of Fusarium graminearum: opportunities and challenges. New Phytol 207:1–3. https://doi.org/10.1111/nph.13457
Jiang J, Yun Y, Liu Y, Ma Z (2012) FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in Fusarium graminearum. Fungal Genet Biol Fg B 49:653–662. https://doi.org/10.1016/j.fgb.2012.06.005
Jiang C, Zhang C, Wu C, Sun P, Hou R, Liu H, Wang C, Xu JR (2016) TRI6 and TRI10 play different roles in the regulation of deoxynivalenol (DON) production by cAMP signalling in Fusarium graminearum. Environ Microbiol 18:3689–3701. https://doi.org/10.1111/1462-2920.13279
Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. https://doi.org/10.1016/S0092-8674(01)00434-2
Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3:893–905. https://doi.org/10.1038/nrm973
Kershaw MJ, Talbot NJ (1998) Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol 23:18–33. https://doi.org/10.1006/fgbi.1997.1022
Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M (2007) Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. J Agric Chem Soc Jpn 71:2105–2123. https://doi.org/10.1271/bbb.70183
Kris A, Adriaan V, Monica H, Geert H (2014) Deoxynivalenol: a major player in the multifaceted response of Fusarium to its environment. Toxins 6:1–19. https://doi.org/10.3390/toxins6010001
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Maxfield FR, Mcgraw TE (2004) Endocytic recycling. Nat Rev Mol Cell Biol 5:121–132. https://doi.org/10.1038/nrm1315
Mcgough IJ, Cullen PJ (2011) Recent advances in retromer biology. Traffic 12:963–971. https://doi.org/10.1111/j.1600-0854.2011.01201.x
Mcmullen M, Bergstrom G, Wolf ED, Dill-Macky R, Hershman D, Shaner G, Sanford DV (2012) A unified effort to fight an enemy of wheat and barley: fusarium head blight. Plant Dis 96:1712–1728. https://doi.org/10.1094/PDIS-03-12-0291-FE
Merrill SA, Hanson PI (2010) Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem 285:35428–35438. https://doi.org/10.1074/jbc.M110.126318
Morita E (2012) Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. Febs J 279:1399–1406. https://doi.org/10.1111/j.1742-4658.2012.08534.x
Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM (2002) Estimating the economic impact of a crop disease: the case of Fusarium Head Blight in the wheat and barley. 2002 National Fusarium Head Blight Forum Proceedings 275–281
Nickerson DP, West M, Odorizzi G (2006) Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol 175:715–720. https://doi.org/10.1083/jcb.200606113
Nickerson DP, West M, Henry R, Odorizzi G (2010) Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell 21:1023–1032. https://doi.org/10.1091/mbc.e09-09-0776
Paez VJ, Goodman K, Otegui MS (2016) Endocytosis and Endosomal trafficking in plants. Annu Rev Plant Biol 67:309–335. https://doi.org/10.1146/annurev-arplant-043015-112242
Pestka JJ, Smolinski AT (2005) Deoxynivalenol: toxicology and potential effects on humans. J Toxicol Environ Health Part B 8:39–69. https://doi.org/10.1080/10937400590889458
Piper RC, Cooper AA, Yang H, Stevens TH (1995) VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol 131:603–617. https://doi.org/10.1083/jcb.131.3.603
Proctor RH, Hohn TM, Mccormick SP (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact 8:593–601. https://doi.org/10.1094/MPMI-8-0593
Raiborg C, Rusten TE, Stenmark H (2003) Protein sorting into multivesicular endosomes. Curr Opin Cell Biol 15:446–455. https://doi.org/10.1016/S0955-0674(03)00080-2
Rue SM, Mattei S, Saksena S, Emr SD (2008) Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell 19:475–484. https://doi.org/10.1091/mbc.e07-07-0694
Saksena S, Sun J, Chu T, Emr SD (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem Sci 32:561–573. https://doi.org/10.1016/j.tibs.2007.09.010
Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346. https://doi.org/10.1007/BF00340712
Schuh AL, Audhya A (2014) The ESCRT machinery: From the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol 49:242–261. https://doi.org/10.3109/10409238.2014.881777
Seong K, Li L, Hou Z, Tracy M, Kistler HC, Xu JR (2006) Cryptic promoter activity in the coding region of the HMG-CoA reductase gene in Fusarium graminearum. Fungal Genet Biol 43:34–41. https://doi.org/10.1016/j.fgb.2005.10.002
Slagsvold T, Marchese A, Brech A, Stenmark H (2006) CISK attenuates degradation of the chemokine receptor CXCR4 via the ubiquitin ligase AIP4. EMBO J 16:3738–3749. https://doi.org/10.1038/sj.emboj.7601267
Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G (2002) The biogenesis and functions of exosomes. Traffic 5:321–330. https://doi.org/10.1034/j.1600-0854.2002.30502.x
Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM (2006) A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics 88:333–346. https://doi.org/10.1016/j.ygeno.2006.04.003
Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792. https://doi.org/10.1083/jcb.128.5.779
Wösten HA, Richter M, Willey JM (1999) Structural proteins involved in emergence of microbial aerial hyphae. Fungal Genet Biol 27:153–160. https://doi.org/10.1006/fgbi.1999.1130
Xie Q, Chen A, Zheng W, Xu H, Shang W, Zheng H, Zhang D, Zhou J, Lu G, Li G (2016) Endosomal sorting complexes required for transport-0 is essential for fungal development and pathogenicity in Fusarium graminearum. Environ Microbiol 18:3742–3757. https://doi.org/10.1111/1462-2920.13296
Xu YB, Li HP, Zhang JB, Bo S, Chen FF, Duan XJ, Xu HQ, Liao YC (2010) Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet Biol 47:205–215. https://doi.org/10.1016/j.fgb.2009.11.003
Yang P, Chen Y, Wu H, Fang W, Liang Q, Zheng Y, Olsson S, Zhang D, Zhou J, Wang Z, Zheng W (2018) The 5-oxoprolinase is required for conidiation, sexual reproduction, virulence and deoxynivalenol production of Fusarium graminearum. Curr Genet 64:285–301. https://doi.org/10.1007/s00294-017-0747-y
Yun Y, Liu Z, Zhang J, Shim WB, Chen Y, Ma Z (2014) The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ Microbiol 16:2023–2037. https://doi.org/10.1111/1462-2920.12334
Zheng W, Xu Z, Xie Q, Huang Q, Zhang C, Zhai H, Xu L, Lu G, Won-Bo S, Wang Z (2012) A conserved homeobox transcription factor Htf1 Is required for phialide development and conidiogenesis in Fusarium species. Plos One 7:e45432. https://doi.org/10.1371/journal.pone.0045432
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31601583 & 31670142), the FAFU international cooperation project (KXB16010A), the FAFU Outstanding Youth Scientific Research Project (xjq201625), the Graduate School Research Foundation (No. 324-1122yb030) and the Plant Protection College Foundation of Fujian Agriculture and Forestry University.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by M. Kupiec.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2019_949_MOESM1_ESM.tif
Supplementary material 1 Identification of homologous ESCRT-III accessory proteins in F. graminearum. A Comparative analysis of orthologs of ESCRT-III accessory proteins between S. cerevisiae and F. graminearum. Alignment of amino acid sequences of B F. graminearum FgVps60 and S. cerevisiae ScVps60, C FgIst1 and ScIst1, D FgDid2 and ScDid2. (TIF 4533 KB)
294_2019_949_MOESM2_ESM.tif
Supplementary material 2 Deletion of FgVPS60, FgIST1 and FgDID2 gene from F. graminearum. FgVPS60, FgIST1 and FgDID2 are marked large white arrows while hph gene is marked with black arrow. Primers used in this study is marked with small and black arrows. A Targeted gene deletion of F. graminearum FgVPS60. Southern blots of KpnI-digested genomic DNA from wild-type strain PH-1 and putative ΔFgvps60 transformants were hybridized with probe marked in left panel. A 2.2-kb band was detected in the wild type, while 2.7-kb bands were detected in the two independent ΔFgvps60 mutants (right panel). B Targeted gene deletion of F. graminearum FgIST1. Southern blots of XhoI-digested genomic DNA from wild-type strain PH-1 and putative ΔFgist1 transformants were hybridized with probe marked in left panel. A 2.9-kb band was detected in the wild type, while 4.2-kb bands were detected in the two independent ΔFgist1 mutants (right panel). C Targeted gene deletion of F. graminearum FgDID2. Southern blots of XhoI-digested genomic DNA from wild-type strain PH-1 and putative ΔFgdid2 transformants were hybridized with probe marked in left panel. A 1.0-kb band was detected in the wild type, while 2.7-kb bands were detected in the two independent ΔFgdid2 mutants (right panel). (TIF 529 KB)
294_2019_949_MOESM3_ESM.tif
Supplementary material 3 Relative expression level of hydrophobin gene FGSG_03960 in PH-1 and the ESCRT-III accessory proteins deletion mutants. RNA samples were extracted from mycelia of the strains after inoculation in liquid CM medium for 3 days. The expression level in wild-type PH-1 was arbitrarily set to 1. Line bars in each column represent the standard deviation (SD) from three independent experiments. *p<0.05; **p<0.01; ***p<0.001; Student’s T test was used. (TIF 194 KB)
294_2019_949_MOESM4_ESM.tif
Supplementary material 4 Ascospores release of PH-1, ΔFgist1 and ΔFgist1-com strains. Fascicles of asci observed by a microscope after squeezing the perithecia 14 days after self-cross of each strain on carrot plates. Scale bar = 50 μm. (TIF 2545 KB)
Rights and permissions
About this article
Cite this article
Xie, Q., Chen, A., Zhang, Y. et al. ESCRT-III accessory proteins regulate fungal development and plant infection in Fusarium graminearum. Curr Genet 65, 1041–1055 (2019). https://doi.org/10.1007/s00294-019-00949-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-019-00949-z