Abstract
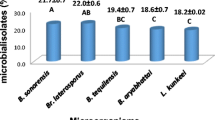
Secondary metabolites of bacteria associated with honey bees were evaluated as part of an investigation on their potentiality for apiary health. Low molecular weight compounds released into culture filtrates by the four bacterial isolates taken from surface of healthy honey bees were analyzed using time-of-flight mass spectrometry. Only one low molecular weight compound was found in the culture filtrate of each isolate. Bacillus thuringiensis, Bifidobacterium asteroides and Acetobacteraceae bacterium, released into culture filtrates platynecine, a pyrrolizidine alkaloid of plant origin, which, until now, had never been reported as produced by bacteria. Lactobacillus kunkeei produced a 3,5-dinitropyridine, of unknown biological action never associated so far to bacteria. The highest relative concentration of platynecine was detected in B. thuringiensis (100%), B. asteroides and A. bacterium showed a concentration above 50% and below 25% that concentration. An in vitro assay on the potential for controlling the parasitic mite Varroa destructor by the culture filtrates of the three platynecine-producing bacteria was performed. Varroa mite mortality was proportional to the platynecine relative concentration into culture filtrate. Although miticidal activity of B. thuringiensis might be associated to other toxic proteins produced by this species, B. asteroides toxicity toward V. destructor along with the other findings of this study support the hypothesis that platynecine plays a direct or an indirect role in controlling varroa. Findings of this study suggest that secondary metabolites released by honey bee-associated bacteria represent a source of natural compounds to be considered in the challenge for counteracting the colony decline.

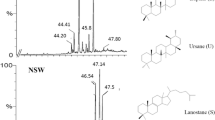
Source: https://www.chemspider.com/Chemical-Structure.83763.html. Features in Table 2

Similar content being viewed by others
References
Smith KM, Loh EH, Rostal MK, Zambrana-Torrelio CM, Mendiola L, Daszak P (2013) Pathogens, pests, and economics: drivers of honey bee colony declines and losses. EcoHealth 10:434–445. https://doi.org/10.1007/s10393-013-0870-2
Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE (2010) Global pollinator declines: trends, impacts and drivers. Trends Ecol Evol 25:345–353. https://doi.org/10.1016/j.tree.2010.01.007
Vanengelsdorp D, Meixner MD (2010) A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol 103(Suppl 1):S80–95. https://doi.org/10.1016/j.jip.2009.06.011
New TR (2016) Alien species and insect conservation. https://doi.org/10.1007/978-3-319-38774-1
Chen YP, Siede R (2007) Honey bee viruses. Adv Virus Res 70:33–80. https://doi.org/10.1016/S0065-3527(07)70002-7
Ramsey SD, Ochoa R, Bauchan G, Gulbronson C, Mowery JD, Cohen A, Lim D, Joklik J, Cicero JM, Ellis JD, Hawthorne D, vanEngelsdorp D (2019) Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc Natl Acad Sci USA 116:1792–1801. https://doi.org/10.1073/pnas.1818371116
Rosenkranz P, Aumeier P, Ziegelmann B (2010) Biology and control of Varroa destructor. J Invertebr Pathol 103(Suppl):S96–119. https://doi.org/10.1016/j.jip.2009.07.016
Currie RW, Pernal SF, Guzmán-Novoa E (2010) Honey bee colony losses in Canada. J Apic Res 49:104–106. https://doi.org/10.3896/IBRA.1.49.1.18
Rademacher E, Imdorf A (2004) Legalization of the use of oxalic acid in varroa control. Bee World 85:70–72. https://doi.org/10.1080/0005772X.2004.11099633
Charrière J-D, Imdorf A (2002) Oxalic acid treatment by trickling against Varroa destructor: recommendations for use in central Europe and under temperate climate conditions. Bee World 83:51–60. https://doi.org/10.1080/0005772X.2002.11099541
Cimmino A, Freda F, Santoro E, Superchi S, Evidente A, Cristofaro M, Masi M (2019) alpha-Costic acid, a plant sesquiterpene with acaricidal activity against Varroa destructor parasitizing the honey bee. Nat Prod Res. https://doi.org/10.1080/14786419.2019.1652291
Shaw KE, Davidson G, Clark SJ, Ball BV, Pell JK, Chandler D, Sunderland KD (2002) Laboratory bioassays to assess the pathogenicity of mitosporic fungi to Varroa destructor (Acari: Mesostigmata), an ectoparasitic mite of the honeybee, Apis mellifera. Biol Control 24:266–276. https://doi.org/10.1016/S1049-9644(02)00029-4
Lodesani M, Costa C, Franceschetti S, Bergomi P, Galaverna G, Dall’Asta C (2017) Toxicity of destruxins against the parasitic mite Varroa destructor and its host Apis mellifera. J Apic Res 56:278–287. https://doi.org/10.1080/00218839.2017.1304611
Meikle WG, Sammataro D, Neumann P, Pflugfelder J (2012) Challenges for developing pathogen-based biopesticides against Varroa destructor (Mesostigmata: Varroidae). Apidologie 43:501–514. https://doi.org/10.1007/s13592-012-0118-0
Tu S, Qiu X, Cao L, Han R, Zhang Y, Liu X (2010) Expression and characterization of the chitinases from Serratia marcescens GEI strain for the control of Varroa destructor, a honey bee parasite. J Invertebr Pathol 104:75–82. https://doi.org/10.1016/j.jip.2010.02.002
Alquisira-Ramírez EV, Peña-Chora G, Hernández-Velázquez VM, Alvear-García A, Arenas-Sosa I, Suarez-Rodríguez R (2017) Effects of Bacillus thuringiensis strains virulent to Varroa destructor on larvae and adults of Apis mellifera. Ecotoxicol Environ Saf 142:69–78. https://doi.org/10.1016/j.ecoenv.2017.03.050
Tsagou V, Lianou A, Lazarakis D, Emmanouel N, Aggelis G (2004) Newly isolated bacterial strains belonging to Bacillaceae (Bacillus sp.) and Micrococcaceae accelerate death of the honey bee mite, Varroa destructor (V. jacobsoni), in laboratory assays. Biotechnol Lett 26:529–532. https://doi.org/10.1023/B:BILE.0000019563.92959.0e
Sabaté DC, Cruz MS, Benitez-Ahrendts MR, Audisio MC (2012) Beneficial effects of Bacillus subtilis subsp. subtilis Mori2, a honey-associated strain, on honeybee colony performance. Probiotics Antimicrobial Proteins 4:39–46. https://doi.org/10.1007/s12602-011-9089-0
Audisio MC, Sabaté DC, Benítez-Ahrendts MR (2015) Effect of Lactobacillus johnsonii CRL1647 on different parameters of honeybee colonies and bacterial populations of the bee gut. Beneficial Microbes 6:687–695. https://doi.org/10.3920/BM2014.0155
Crotti E, Balloi A, Hamdi C, Sansonno L, Marzorati M, Gonella E, Favia G, Cherif A, Bandi C, Alma A, Daffonchio D (2012) Microbial symbionts: a resource for the management of insect-related problems. Microb Biotechnol 5:307–317. https://doi.org/10.1111/j.1751-7915.2011.00312.x
Olofsson TC, Butler E, Markowicz P, Lindholm C, Larsson L, Vasquez A (2014) Lactic acid bacterial symbionts in honeybees—an unknown key to honey’s antimicrobial and therapeutic activities. Int Wound J 13:668–679. https://doi.org/10.1111/iwj.12345
Beemelmanns C, Guo H, Rischer M, Poulsen M (2016) Natural products from microbes associated with insects. Beilstein J Org Chem 12:314–327
Crawford JM, Clardy J (2011) Bacterial symbionts and natural products. Chem Commun (Camb) 47:7559–7566. https://doi.org/10.1039/c1cc11574j
Schmidt EW (2008) Trading molecules and tracking targets in symbiotic interactions. Nat Chem Biol 4:466–473. https://doi.org/10.1038/nchembio.101
Endo A, Salminen S (2013) Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst Appl Microbiol 36:444–448. https://doi.org/10.1016/j.syapm.2013.06.002
Rampersad J, Khan A, Ammons D (2002) Usefulness of staining parasporal bodies when screening for Bacillus thuringiensis. J Invertebr Pathol 79:203–204. https://doi.org/10.1016/S0022-2011(02)00018-6
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. https://doi.org/10.1093/nar/gkv951
Kleinbaum DG, Klein M (2012) Survival analysis. A self-learning text, 3rd edn. Springer-Verlag, New York. https://doi.org/10.1007/978-1-4419-6646-9
Moreira R, Pereira MD, Valentão P, Andrade BP (2018) Pyrrolizidine alkaloids: chemistry, pharmacology, toxicology and food safety. Int J Mol Sci. https://doi.org/10.3390/ijms19061668
Winkler M, Cakir B, Sander W (2004) 3,5-Pyridyne—a heterocyclic meta-benzyne derivative. J Am Chem Soc 126:6135–6149. https://doi.org/10.1021/ja039142u
Schoning V, Hammann F, Peinl M, Drewe J (2017) Editor’s highlight: identification of any structure-specific hepatotoxic potential of different pyrrolizidine alkaloids using random forests and artificial neural networks. Toxicol Sci 160:361–370. https://doi.org/10.1093/toxsci/kfx187
Hartmann T, Ober D (2000) Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. In: Leeper FJ, Vederas JC (eds) Biosynthesis—aromatic polyketides, isoprenoids, alkaloids. Springer, Berlin, pp 207–243
Ober D (2003) Chemical ecology of alkaloids exemplified with the pyrrolizidines. In: Romeo JTBT-RA in P (ed) Integrative phytochemistry: from ethnobotany to molecular ecology. Elsevier, Amsterdam, pp 203–230
Schimming O, Challinor VL, Tobias NJ, Adihou H, Grun P, Poschel L, Richter C, Schwalbe H, Bode HB (2015) Structure, biosynthesis, and occurrence of bacterial pyrrolizidine alkaloids. Angew Chem (International ed in English) 54:12702–12705. https://doi.org/10.1002/anie.201504877
Joosten L, van Veen JA (2011) Defensive properties of pyrrolizidine alkaloids against microorganisms. Phytochem Rev 10:127–136. https://doi.org/10.1007/s11101-010-9204-y
Ruan J, Liao C, Ye Y, Lin G (2014) Lack of metabolic activation and predominant formation of an excreted metabolite of nontoxic platynecine-type pyrrolizidine alkaloids. Chem Res Toxicol 27:7–16. https://doi.org/10.1021/tx4004159
Palma L, Muñoz D, Berry C, Murillo J, Caballero P (2014) Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6:3296–3325. https://doi.org/10.3390/toxins6123296
Schmidt R, Ulanova D, Wick LY, Bode HB, Garbeva P (2019) Microbe-driven chemical ecology: past, present and future. ISME J 13:2656–2663. https://doi.org/10.1038/s41396-019-0469-x
Vuong HQ, McFrederick QS (2019) Comparative genomics of wild bee and flower isolated lactobacillus reveals potential adaptation to the bee host. Genome Biol Evol 11:2151–2161. https://doi.org/10.1093/gbe/evz136
Sun Z, Zhang W, Guo C, Yang X, Liu W, Wu Y, Song Y, Kwok LY, Cui Y, Menghe B, Yang R, Hu L, Zhang H (2015) Comparative genomic analysis of 45 type strains of the genus Bifidobacterium: a snapshot of its genetic diversity and evolution. PLoS ONE 10:e0117912–e0117912. https://doi.org/10.1371/journal.pone.0117912
Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SGE, Vasquez A (2015) Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16:284. https://doi.org/10.1186/s12864-015-1476-6
Gil MI, Ferreres F, Ortiz A, Subra E, Tomas-Barberan FA (1995) Plant phenolic metabolites and floral origin of rosemary honey. J Agric Food Chem 43:2833–2838. https://doi.org/10.1021/jf00059a012
Ferreres F, García-Viguera C, Tomás-Lorente F, Tomás-Barberán FA (1993) Hesperetin: a marker of the floral origin of citrus honey. J Sci Food Agric 61:121–123. https://doi.org/10.1002/jsfa.2740610119
Truchado P, Martos I, Bortolotti L, Sabatini AG, Ferreres F, Tomas-Barberan FA (2009) Use of quinoline alkaloids as markers of the floral origin of chestnut honey. J Agric Food Chem 57:5680–5686. https://doi.org/10.1021/jf900766v
Baracchi D, Brown MJF, Chittka L (2015) Behavioural evidence for self-medication in bumblebees? F1000Research 4:73. https://doi.org/10.12688/f1000research.6262.2
Negri P, Villalobos E, Szawarski N, Damiani N, Gende L, Garrido M, Maggi M, Quintana S, Lamattina L, Eguaras M (2019) Towards precision nutrition: a novel concept linking phytochemicals, immune response and honey bee health. Insects. https://doi.org/10.3390/insects10110401
Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, Cornman RS, Dainat J, de Miranda JR, Doublet V, Emery O, Evans JD, Farinelli L, Flenniken ML, Granberg F, Grasis JA, Gauthier L, Hayer J, Koch H, Kocher S, Martinson VG, Moran N, Munoz-Torres M, Newton I, Paxton RJ, Powell E, Sadd BM, Schmid-Hempel P, Schmid-Hempel R, Song SJ, Schwarz RS, vanEngelsdorp D, Dainat B (2016) The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio. https://doi.org/10.1128/mBio.02164-15
Price-Whelan A, Dietrich LEP, Newman DK (2006) Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78. https://doi.org/10.1038/nchembio764
Davies J, Ryan KS (2012) Introducing the parvome: bioactive compounds in the microbial world. ACS Chem Biol 7:252–259. https://doi.org/10.1021/cb200337h
Acknowledgements
We would like to thank Dr. Rita Resca of Renolab srl and her staff for the analysis of bacterial metabolites. This study was performed within the frame of CREA research facilities by using institutional resources, and partially supported by EU Reg. 1308/2013 of the Italian Ministry of Agriculture 2016–2019.
Author information
Authors and Affiliations
Contributions
MLM: investigation-secondary metabolites; data analysis; writing—original draft. SML: investigation and methodology-bacteria, writing—review & editing. LM: investigation-methodology-honey bees and Varroa.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Manici, L.M., Saccà, M.L. & Lodesani, M. Secondary Metabolites Produced by Honey Bee-Associated Bacteria for Apiary Health: Potential Activity of Platynecine. Curr Microbiol 77, 3441–3449 (2020). https://doi.org/10.1007/s00284-020-02153-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02153-6