Abstract
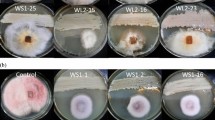
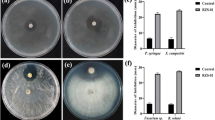
The current study was designed to isolate, identify and characterize a Bacillus sp. capable of producing protease and exhibiting antifungal activity. A highly potent bacterium capable of producing protease abundantly was isolated from the soil collected from the waste pit near Microbiology Laboratory of Birendra Multiple Campus, Bharatpur and later on identified as Lysinibacillus fusiformis strain SK on the basis of morphological, physiological, biochemical and 16S rDNA gene sequencing techniques. The strain SK showed 98.36% similarity with L. fusiformis strain NBRC 15717. Using R-programming statistical analysis tool, the optimum incubation time for the highest average protease production (APP) (47.2 U/mL) was found to be 22 h at 50 °C and both incubation time and temperature showed a significant impact on the production of protease (P < 0.01). The maximum average relative protease activity (ARPA) was observed at pH 7.8 and 48 °C, whereas the least ARPA was observed in the presence of 80 g/L NaCl and 10 g/L HgCl2 (P < 0.01). The newly isolated bacterial strain also exhibited strong antifungal activity against aflatoxigenic Aspergillus flavus and Aspergillus parasiticus suggesting that it can be a potential candidate for protease production and activity over a wider range of temperature and pH as well as for synthesizing effective antifungal compounds.





Similar content being viewed by others
References
Anwar A, Saleemuddin M (1998) Alkaline proteases: a review. Bioresour Technol 64:175–183. https://doi.org/10.1016/S0960-8524(97)00182-X
Sumantha A, Larroche C, Pandey A (2006) Microbiology and industrial biotechnology of food-grade. Food Technol Biotechnol 44:211–220
Agasthya AS, Sharma N, Mohan A, Mahal P (2013) Isolation and molecular characterisation of alkaline protease producing Bacillus thuringiensis. Cell Biochem Biophys 66:45–51. https://doi.org/10.1007/s12013-012-9396-4
Dickinson L (2017) Global proteases market—growth, trends and forecasts (2017–2022). https://www.researchandmarkets.com/reports/4115040/global-proteases-market-growth-trends-and. Accessed 9 Aug 2019
Sandhya C, Sumantha A, Pandey A (2004) Proteases. In: Pandey A, Webb C, Soccol CR, Larroche C (eds) Enzyme technology. Asiatech Publishers Inc., New Delhi, pp 312–325
Rai SK, Roy JK, Mukherjee AK (2010) Characterisation of a detergent-stable alkaline protease from a novel thermophilic strain Paenibacillus tezpurensis sp. nov. AS-S24-II. Appl Microbiol Biotechnol 85:1437–1450. https://doi.org/10.1007/s00253-009-2145-y
Suberu Y, Akande I, Samuel T, Lawal A, Olaniran A (2019) Optimization of protease production in indigenous Bacillus species isolated from soil samples in Lagos, Nigeria using response surface methodology. Biocatal Agric Biotechnol. https://doi.org/10.1016/j.bcab.2019.01.049
Outtrup HB, Dambmann CS, Lindegaard P (1994) Proteases (United States Patent). 1–8
Lawler D, Smith S (2001) Enzyme-producing strain of Bacillus bacteria (Patent). 1–5
Cordeiro CAM, Martins MLL, Luciano AB (2004) Production and properties of an α-amylase from thermophilic Bacillus sp. Braz J Microbiol 35:91–96. https://doi.org/10.1590/s1517-83822004000100015
Bergey DH, Vos PD, Whitman WB (2009) Bergey’s manual of systematic bacteriology. In: Whitman WB (ed) The Firmicutes, 2nd edn. Springer, Dordrecht
Ahmed I, Yokota A, Yamazoe A, Fujiwara T (2007) Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int J Syst Evol Microbiol 57:1117–1125. https://doi.org/10.1099/ijs.0.63867-0
Mechri S, Kriaa M, Ben Elhoul Berrouina M, Omrane Benmrad M, Zaraî Jaouadi N, Rekik H, Bouacem K, Bouanane-Darenfed A, Chebbi A, Sayadi S, Chamkha M, Bejar S, Jaouadi B (2017) Optimized production and characterization of a detergent-stable protease from Lysinibacillus fusiformis C250R. Int J Biol Macromol 101:383–397. https://doi.org/10.1016/j.ijbiomac.2017.03.051
Nshimiyimana JB, Khadka S, Mwizerwa EM, Akimana N, Adhikari S, Nsabimana A (2018) Thermophiles: isolation, characterization and screening for enzymatic activity. Biosci Discov 9:430–437
Sapkota S, Khadka S, Gautam A, MaharjanR SR, Dhakal S, Panta OP, Khanal S, Poudel P (2019) Screening and optimization of thermo-tolerant Bacillus sp. for amylase production and antifungal activity. J Inst Sci Technol 24:47–56. https://doi.org/10.3126/jist.v24i1.24628
Che J, Liu B, Liu G, Che J, Bo L, Liu G, Chen Q, Lan J (2017) Volatile organic compounds produced by Lysinibacillus sp. FJAT-4748 possess antifungal activity against Colletotrichum acutatum. Biocontrol Sci Technol 27:1349–1362. https://doi.org/10.1080/09583157.2017.1397600
Yu J, Payne GA, Nierman WC, Machida M, Bennett JW, Campbell BC, Robens JF, Bhatnagar D, Dean RA, Cleveland TE (2008) Aspergillus flavus genomics as a tool for studying the mechanism of aflatoxin formation. Food Addit Contam A 25:1152–1157. https://doi.org/10.1080/02652030802213375
Cleveland TE, Yu J, Fedorova N, BhatnagarD PayneGA, NiermanWC BJW (2009) Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol 27:151–157. https://doi.org/10.1016/j.tibtech.2008.11.008
Zain ME (2011) Impact of mycotoxins on humans and animals. J Saudi Chem Soc 15:129–144. https://doi.org/10.1016/j.jscs.2010.06.006
Petzinger E, Ziegler K (2000) Ochratoxin A from a toxicological perspective. J Vet Pharmacol Ther 23:91–98. https://doi.org/10.1046/j.1365-2885.2000.00244.x
FAO (2001) Manual on the application of the HACCP system in mycotoxin prevention and control. In: FAO food and nutrition paper https://www.fao.org/3/y1390e/y1390e00.htm
Fratamico PM, Bhunia AK, Smith JL (2008) Foodborne pathogens: microbiology and molecular biology. Horizon Scientific Press, Norofolk
Gourama H, Bullerman LB (2016) Aspergillus flavus and Aspergillus parasiticus: aflatoxigenic fungi of concern in foods and feeds: a review. J Food Prot 58:1395–1404. https://doi.org/10.4315/0362-028x-58.12.1395
International Agency for Research on Cancer (IARC) (2002) Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC monographs on the evaluation of carcinogenic risks to hummans. International Agency for Research on Cancer, Lyon
Liu Y, Wu F (2010) Global burden of Aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ Health Perspect 118:818–824. https://doi.org/10.1289/ehp.0901388
Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW (2007) Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677–1692. https://doi.org/10.1099/mic.0.2007/007641-0
Petchkongkaew A, Taillandier P, Gasaluck P, Lebrihi A (2008) Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): screening for aflatoxin B1 and ochratoxin a detoxification. J Appl Microbiol 104:1495–1502. https://doi.org/10.1111/j.1365-2672.2007.03700.x
Pitt JI, Wild CP, Baan RA, Gelderblom WCA, Miller JD, Rile RT, Wu F (2012) Improving public health through mycotoxin control. Lyon, France: (IARC Scientific Publications Series, No. 158).
World Health Organization (WHO) (2018) Food safety and digest. In: Department of food safety and zoonoses. https://www.scopus.com/inward/record.uri?eid=2-s2.0-0034270430&partnerID=40&md5=1cfd91de5ef6da00ed45d7b745f4bd6b. Accessed 10 Aug 2019
CDC (2004) Outbreak of aflatoxin poisoning—eastern and central provinces, Kenya. Centers for disease control and prevention. MMWR 53:790–793
Kumar S, Yadav B, Premarajan K, Koirala P (2005) Occurrence of aflatoxin in some of the food and feed in Nepal. Indian J Med Sci 59:331. https://doi.org/10.4103/0019-5359.16649
Desjardins AE, Plattner RD, Maragos CM, McCormick SP, Manandhar G, Shrestha K (2000) Occurrence of Fusarium species and mycotoxins in Nepalese maize and wheat and the effect of traditional processing methods on mycotoxin levels. J Agric Food Chem 48:1377–1383. https://doi.org/10.1021/jf991022b
Galil OAA (1992) Fermentation of proteases by Aspergillus fumigates and Pencillium sp. J King Saud Univ 4:127–136
Krieg NR, Holt JC (1984) Bergey’s manual of systematic bacteriology, 1st edn. Williams and Wilkins, Baltimore
Miaoying C (2001) East show strains, Cai Miaoying common bacterial identification manual system. Science Press, Beijing
Forbes B, Sahm D, Weissfeld A (2007) Bailey & Scott’s diagnostic microbiology, 12th edn. Mosby, Saint Louis
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Clinical and Laboratory Standards Institute (CLSI) (2017) Performance standards for antimicrobial susceptibility testing. In: CLSI supplement M100, 27th edn. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania, 19087, USA
Vijayaraghavan P, Gnana S, Vincent P (2013) A simple method for the detection of protease activity on agar plates using bromocresolgreen dye. J Biochem Technol 4:628–630
Tekin N, Cihan AÇ, Takaç ZS, Tüzün CY, Tunç K, Çökmüş C (2012) Alkaline protease production of Bacillus cohnii APT5. Turk J Biol 36:430–440
Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54:109–114
Bacon CW, Hinton DM (2002) Endophytic and biological control potential of Bacillus mojavensis and related species. Biol Control 23:274–284
Miwa H, Ahmed I, Yokota A, Fujiwara T (2009) Lysinibacillus parviboronicapiens sp. nov., a low-boron-containing bacterium isolated from soil. Int J Syst Evol Microbiol 59:1427–1432. https://doi.org/10.1099/ijs.0.65455-0
Priest FG, Goodfellow M, Todd C (1988) A numerical classification of the genus Bacillus. Microbiology 134:1847–1882. https://doi.org/10.1099/00221287-134-7-1847
Wang L, Fan D, Chen W, Terentjev EM (2015) Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci Rep 5:1–11. https://doi.org/10.1038/srep15159
Hui C, Wei R, Jiang H, Zhao Y, Xu L (2019) Characterization of the ammonification, the relevant protease production and activity in a high-efficiency ammonifier Bacillus amyloliquefaciens DT. Int Biodeterior Biodegrad 142:11–17. https://doi.org/10.1016/j.ibiod.2019.04.009
Margesin R, Schinner F (1992) Extracellular protease production by psychrotrophic bacteria from glaciers. Int Biodeterior Biodegrad 29:177–189
Dudhgara PR, Sunil B, Anjana G (2015) Hide dehairing and laundry detergent compatibility testing of thermostable and solvents tolerant alkaline protease from hot spring isolate Bacillus cohniiU3. Online J Biol Sci 15:152–161. https://doi.org/10.3844/ojbsci.2015.152.161
Gonçalves RN, Gozzini Barbosa SD, Silva-López RE (2016) Proteases from Canavalia ensiformis: active and thermostable enzymes with potential of application in biotechnology. Biotechnol Res Int 2016:1–11. https://doi.org/10.1155/2016/3427098
Vogt G, Woell S, Argos P (1997) Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol 269:631–643. https://doi.org/10.1006/jmbi.1997.1042
Su NW, Wang ML, Kwok KF, Lee MH (2005) Effects of temperature and sodium chloride concentration on the activities of proteases and amylases in soy sauce koji. J Agric Food Chem 53:1521–1525. https://doi.org/10.1021/jf0486390
Mohan M, Kozhithodi S, Nayarisseri A, Elyas KK (2018) Screening, purification and characterization of protease inhibitor from Capsicum frutescens. Bioinformation 14:285–293. https://doi.org/10.6026/97320630014285
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. https://doi.org/10.1016/j.tim.2007.12.009
Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R (2009) Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140:27–37. https://doi.org/10.1016/j.jbiotec.2008.10.011
Zhao Z, Wang Q, Wang K, BrianK LC, Gu Y (2010) Study of the antifungal activity of Bacillus vallismortis ZZ185 in vitro and identification of its antifungal components. Bioresour Technol 101:292–297. https://doi.org/10.1016/j.biortech.2009.07.071
Ono M, Kimura N (1991) Antifungal peptides produced by Bacillus subtilis for the biological control of aflatoxin contamination. Proc Jpn Assoc Mycotoxicol 34:23–28
Klich MA, Arthur KS, Lax AR, Bland JM (1994) Iturin A: a potential new fungicide for stored grains Maren. Mycopathologia 127:123–127
Moyne AL, Shelby R, Cleveland TE, Tuzun S (2001) Bacillomycin D: an iturin with antifungal activity against Aspergillus flavus. J Appl Microbiol 90:622–629. https://doi.org/10.1046/j.1365-2672.2001.01290.x
Quentin MJ, Besson F, Peypoux F, Michel G (1982) Action of peptidolipidic antibiotics of the iturin group on erythrocytes. Effect of some lipids on hemolysis. BBA 684:207–211. https://doi.org/10.1016/0005-2736(82)90007-4
Funding
No specific funding was provided to conduct this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors state there is no conflict of interests regarding the publication of this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khadka, S., Adhikari, S., Thapa, A. et al. Screening and Optimization of Newly Isolated Thermotolerant Lysinibacillus fusiformis Strain SK for Protease and Antifungal Activity. Curr Microbiol 77, 1558–1568 (2020). https://doi.org/10.1007/s00284-020-01976-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01976-7