Abstract
Background
The immune system has been implicated in the pathophysiology of cutaneous squamous cell carcinoma (cSCC) as evidenced by the substantially increased risk of cSCC in immunosuppressed individuals. Associations between cSCC risk and single nucleotide polymorphisms (SNPs) in the HLA region have been identified by genome-wide association studies (GWAS). The translation of the associated HLA SNPs to structural amino acids changes in HLA molecules has not been previously elucidated.
Methods
Using data from a GWAS that included 7238 cSCC cases and 56,961 controls of non-Hispanic white ancestry, we imputed classical alleles and corresponding amino acid changes in HLA genes. Logistic regression models were used to examine associations between cSCC risk and genotyped or imputed SNPs, classical HLA alleles, and amino acid changes.
Results
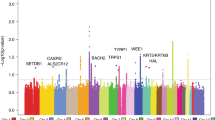
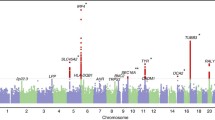
Among the genotyped SNPs, cSCC risk was associated with rs28535317 (OR = 1.20, p = 9.88 × 10− 11) corresponding to an amino-acid change from phenylalanine to leucine at codon 26 of HLA-DRB1 (OR = 1.17, p = 2.48 × 10− 10). An additional independent association was observed for a threonine to isoleucine change at codon 107 of HLA-DQA1 (OR = 1.14, p = 2.34 × 10− 9). Among the classical HLA alleles, cSCC was associated with DRB1*01 (OR = 1.18, p = 5.86 × 10− 10). Conditional analyses revealed additional independent cSCC associations with DQA1*05:01 and DQA1*05:05. Extended haplotype analysis was used to complement the imputed haplotypes, which identified three extended haplotypes in the HLA-DR and HLA-DQ regions.
Conclusions
Associations with specific HLA-DR and -DQ alleles are likely to explain previously observed GWAS signals in the HLA region associated with cSCC risk.




Similar content being viewed by others
Abbreviations
- cSCC:
-
Cutaneous squamous cell carcinoma
- GERA:
-
Genetic Epidemiology Research in Adult Health and Aging
- GWAS:
-
Genome-wide association study
- HLA:
-
Human leukocyte antigen
- KPNC:
-
Kaiser Permanente Northern California
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
References
Jennings L, Schmults CD (2010) Management of high-risk cutaneous squamous cell carcinoma. J Clin Aesthet Dermatol 3(4):39–48
Lindström LS, Yip B, Lichtenstein P et al (2007) Etiology of familial aggregation in melanoma and squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers Prev 16(8):1639–1643
Trakatelli M, Ulrich C, del Marmol V et al (2007) Epidemiology of nonmelanoma skin cancer (NMSC) in Europe: accurate and comparable data are needed for effective public health monitoring and interventions. Br J Dermatol 156(Suppl 3):1–7
Euvrard S, Kanitakis J, Claudy A (2003) Skin cancers after organ transplantation. N Engl J Med 348(17):1681–1691
Silverberg MJ, Leyden W, Warton EM et al (2013) HIV infection status, immunodeficiency, and the incidence of non-melanoma skin cancer. J Natl Cancer Inst 105(5):350 – 60
Asgari MM, Wang W, Ioannidis NM et al (2016) Identification of susceptibility loci for cutaneous squamous cell carcinoma. J Invest Dermatol 136(5):930–937
Chahal HS, Lin Y, Ransohoff KJ et al (2016) Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nat Commun 7:12048
Jia X, Han B, Onengut-Gumuscu S et al (2013) Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8(6):e64683
Hoffmann TJ, Zhan Y, Kvale MN et al (2011) Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics 98(6):422–430
Hoffmann TJ, Kvale MN, Hesselson SE et al (2011) Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics 98(2):79–89
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2(12):e190
Jorgenson E, Melles RB, Hoffmann TJ et al (2016) Common coding variants in the HLA-DQB1 region confer susceptibility to age-related macular degeneration. Eur J Hum Genet 24(7):1049–1055
Chang CC, Chow CC, Tellier LC et al (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7
Ollila HM, Ravel JM, Han F et al (2015) HLA-DPB1 and HLA class I confer risk of and protection from narcolepsy. Am J Hum Genet 96(1):136–46
Raj P, Rai E, Song R et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife 2016;5:e12089, (PMCID: PMC4811771)
Glover MT, Bodmer J, Bodmer W et al (1993) HLA antigen frequencies in renal transplant recipients and non-immunosuppressed patients with non-melanoma skin cancer. Eur J Cancer 29A(4):520–524
Cerimele D, Contu L, Carcassi C et al (1988) HLA and multiple skin carcinomas. Dermatologica 176(4):176–81
Czarnecki DB, Lewis A, Nicholson I et al (1991) Multiple nonmelanoma skin cancer associated with HLA DR7 in southern Australia. Cancer 68(2):439–40
Schreiber RD, Old LJ, Smyth MJ (2011) Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331(6024):1565–1570
Vinay DS, Ryan EP, Pawelec G et al (2015) Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol 35 S185-98
Baecher-Allan C, Wolf E, Hafler DA (2006) MHC class II expression identifies functionally distinct human regulatory T cells. J Immunol 176(8):4622–4631
Hu JM, Li L, Chen YZ, et al. HLA-DRB1 and HLA-DQB1 methylation changes promote the occurrence and progression of Kazakh ESCC. Epigenetics 2014;9(10):1366–1373
Jones EY, Fugger L, Strominger JL et al (2006) MHC class II proteins and disease: a structural perspective. Nat Rev Immunol 6(4):271 – 82
Raychaudhuri S, Sandor C, Stahl EA et al (2012) Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 44(3):291–296
Feitsma AL, van der Helm-van Mil AH, Huizinga TW et al (2008) Protection against rheumatoid arthritis by HLA: nature and nurture. Ann Rheum Dis 67(Suppl 3):iii61-3
Fernandez CA, Smith C, Yang W et al (2014) HLA-DRB1*07:01 is associated with a higher risk of asparaginase allergies. Blood 124(8):1266–1276
Kvale MN, Hesselson S, Hoffmann TJ et al (2015) Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA). Cohort Genetics 200(4):1051–1060
Pereyra F, Jia X, McLaren PJ et al (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330(6010):1551–1557
Butler MO, Ansén S, Tanaka M et al (2010) A panel of human cell-based artificial APC enables the expansion of long-lived antigen-specific CD4 + T cells restricted by prevalent HLA-DR alleles. Int Immunol 22(11):863–73
Funding
This work was supported by a grant from the National Cancer Institute at the National Institutes of Health (Grant number R01CA166672).
Author information
Authors and Affiliations
Contributions
WW: drafting and revisions to manuscript, acquisition of data, and/or analysis and interpretation of results. HMO: drafting and revisions to manuscript, acquisition of data, and/or analysis and interpretation of results. ASW: conception and design, revisions to manuscript. SD: interpretation of results and revisions to manuscript. NMI: revisions to manuscsript. EJ: interpretation of results and revisions to manuscript. EM: conception and design, data analysis, revisions to manuscript. MMA: conception, funding, data acquisition, revisions to manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Dr. Asgari has received institutional research funding (role: Principal Investigator) from Valeant Pharmaceuticals for an unrelated topic (atopic dermatitis). All other authors declare that they have no conflict of interest.
Ethical approval and ethical standards
This study was approved by the Kaiser Permanente Northern California Institutional review board (approval number CN-12MAsga-03-H) and was conducted in accordance with the Helsinki declaration.
Informed consent
Informed consent was waived.
Additional information
Emmanuel Mignot and Maryam M. Asgari equally contributing as last author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, W., Ollila, H.M., Whittemore, A.S. et al. Genetic variants in the HLA class II region associated with risk of cutaneous squamous cell carcinoma. Cancer Immunol Immunother 67, 1123–1133 (2018). https://doi.org/10.1007/s00262-018-2168-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-018-2168-2