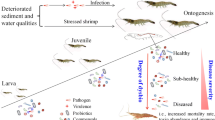
Abstract
Ample evidence shows dysbiosis in the gut microbiota when comparing healthy shrimp with those affected by severe acute hepatopancreatic necrosis disease (AHPND). However, the static comparison used in available studies leads to the uncertainties regarding how and to what extent the gut microbiota responds to the progressive severity of AHPND. In addition, shrimp AHPND is featured by rapid and massive mortality, thus the initiation of AHPND must be diagnosed for preemptive therapy. For these reasons, we explored the ecological assembly of gut microbiota over shrimp AHPND progression. Increasing AHPND severity was associated with linear increase in the copies of pirAB genes, relative abundance of gut Vibrio and potentially pathogenic, and reduction in the gut bacterial diversity, stability, and relative abundance of Bdellovibrio. Negative and significant association between gut Vibrio and Bdellovibrio were noted, indicating that compromised predation exerts a role in AHPND progression. Notably, the extents of departure to the healthy shrimp gut microbiota were positively coupled with the increasing severity of AHPND. After controlling the temporal variation in the gut microbiota as healthy shrimp age, we constructed a diagnosis model that accurately diagnosed the initial, progressed or moribund stages of AHPND, with an overall accuracy of 86.5%. Shrimp AHPND induced more stochastic gut microbiotas as a consequence of the attenuated ability of diseased shrimp to select their commensals, resulting in convergent bacterial communities between gut and rearing water over AHPND progression. Collectively, our findings provide important step toward the ecological assembly of gut microbiota implicating in AHPND etiology and in diagnosing AHPND stages.
Key points
• The departure of shrimp gut microbiota positively linked with AHPND severity.
• The diagnosis model accurately diagnosed the stages of AHPND.
• Shrimp AHPND induced more stochastic gut microbiota.






Similar content being viewed by others
Data availability
Raw sequence data were deposited into the BIG Data Center, CAS under code CRA011876 and are available at the following URL: https://ngdc.cncb.ac.cn/search/?dbId=&q=CRA011876.
References
Anderson MJ, Willis TJ (2003) Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84:511–525
Angthong P, Uengwetwanit T, Uawisetwathana U, Koehorst JJ, Arayamethakorn S, Schaap PJ, Dos Santos VM, Phromson M, Karoonuthaisiri N, Chaiyapechara S, Rungrassamee W (2023) Investigating host-gut microbial relationship in Penaeus monodon upon exposure to Vibrio harveyi. Aquaculture 567:739252
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Browne MW (2000) Cross-Validation Methods J Math Psychol 44:108–132
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Chaiyapechara S, Uengwetwanit T, Arayamethakorn S, Bunphimpapha P, Rungrassamee W (2022) Understanding the host-microbe-environment interactions: intestinal microbiota and transcriptomes of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture 546:737371
Chen WY, Ng TH, Wu JH, Chen JW, Wang HC (2017) Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancre. Sci Rep 7:9395
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M, Ochoa-Romo JP, Ochoa-Leyva A (2018) A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ 6:e5382
Dai W, Sheng Z, Chen J, Xiong J (2020) Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture 517:734802
Donaldson G, Lee SM, Mazmanian SK (2016) Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32
Duong ND, Mai-Hoang TD, Nguyen-Phuoc KH, Do KT, Nguyen NT, Tran TL, Tran-Van H (2023) Monitoring the secreted profile of PirA(vp) and PirB(vp) toxins from Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease. Aquac Int 31:1677–1684
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
El-Saadony MT, Shehata AM, Alagawany M, Abdel-Moneim AE, Selim DA, Abdo M, Khafaga AF, El-Tarabily KA, El-Shall NA, Abd El-Hack ME (2022) A review of shrimp aquaculture and factors affecting the gut microbiome. Aquac Int 30:2847–2869
Giatsis C, Sipkema D, Smidt H, Verreth J, Verdegem M (2014) The colonization dynamics of the gut microbiota in tilapia larvae. PLoS ONE 9:e103641
Holt CC, Bass D, Stentiford GD, Giezen M (2022) Understanding the role of the shrimp gut microbiome in health and disease. J Invertebr Pathol 186:107387
Hossain MS, Dai J, Qiu D (2021) Dysbiosis of the shrimp (Penaeus monodon) gut microbiome with AHPND outbreaks revealed by 16S rRNA metagenomics analysis. Aquac Res 52:3336–3349
Hou D, Huang Z, Zeng S, Liu J, Wei D, Deng X, Weng S, Yan Q, He J (2018) Intestinal bacterial signatures of white feces syndrome in shrimp. Appl Microbiol Biotechnol 102:3701–3709
Huang Z, Zeng S, Xiong J, Hou D, Zhou R, Xing C, Wei D, Deng X, Yu L, Wang H, Deng Z, Weng S, Kriengkrai S, Ning D, Zhou J, He J (2020) Microecological Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8:32
Jose PA, Maharshi A, Jha B (2021) Actinobacteria in natural products research: Progress and prospects. Microbiol Res 246:126708
Kongrueng J, Mitraparp-Arthorn P, Bangpanwimon K, Robins W, Vuddhakul V, Mekalanos J (2017) Isolation of Bdellovibrio and like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Dis Aquatic Organ 124:223–232
Kumar R, Ng TH, Wang HC (2020) Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev Aquac 12:1867–1880
Lavrinienko A, Tukalenko E, Kesniemi J, Kivisaari K, Masiuk S, Boratyński Z, Mousseau TA, Milinevsky G, Mappes T, Watts PC (2020) Applying the Anna Karenina principle for wild animal gut microbiota: temporal stability of the bank vole gut microbiota in a disturbed environment. J Animal Ecol 89:2617–2630
Lee CT, Chen IT, Yang YT, Ko TP, Huang Y, Huang J, Huang M, Lin S, Chen C, Lin S, Lightner D, Wang H, Wang A, Wang H, Hor L, Lo C (2015) The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Nat Acad Sci USA 112:10798–10803
Li E, Xu C, Wang X, Wang S, Zhao Q, Zhang M, Qin JG, Chen L (2018) Gut microbiota and its modulation for healthy farming of Pacific white shrimp Litopenaeus vannamei. Rev Fish Sci Aquac 26:381–399
Liao M, Long X, He Z, Zhao J, Chen X, Zhu D, Sun C (2022) The effect of “Fishery-PV Integration” on Penaeus monodon culture and research on the micro-ecological environment. Front Mar Sci 9:963331
Limberger R, Daugaard U, Gupta A, Krug RM, Lemmen KD, van Moorsel SJ, Suleiman M, Zuppinger-Dingley D, Petchey OL (2023) Functional diversity can facilitate the collapse of an undesirable ecosystem state. Ecol Lett 26:883–895
Lu J, Zhang X, Qiu Q, Chen J, Xiong J (2020) Identifying potential polymicrobial pathogens: Moving beyond differential abundance to driver taxa. Microb Ecol 80:447–458
Lu J, Li X, Qiu Q, Chen J, Xiong J (2022) Gut interkingdom predator-prey interactions are key determinants of shrimp health. Aquaculture 546:737304
Mallon CA, Elsas JV, Salles JF (2015) Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol 23:719–729
Mao J, Lu J, Chen J, Xiong J (2023) Consistent features of the gut microbiota in response to diverse shrimp Litopenaeus vannamei diseases: A meta-analysis. Fish Fish. https://doi.org/10.1111/faf.12787
Ning D, Yuan M, Wu L, Zhang Y, Zhou J (2020) A quantitative framework reveals the ecological drivers of grassland soil microbial community assembly in response to warming. Nat Commun 11:4717
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Core Team (2018) R: A language and environment for statistical computing, reference index version 4.0.2. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. Accessed 22 May 2020
Roughgarden J (2023) Holobiont evolution: Population genetic theory for the hologenome. Am Nat 201:763–778
Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N (2016) Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J Invertebr Pathol 133:12–19
Sha H, Lu J, Chen J, Xiong J (2022) A meta-analysis study of the robustness and universality of gut microbiota-shrimp diseases relationship. Environ Microbiol 24:3924–3938
Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JH (2012) Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417
Shen H, Song T, Lu J, Qiu Q, Chen J, Xiong J (2021) Shrimp AHPND causing Vibrio anguillarum infection: Quantitative diagnosis and identifying antagonistic bacteria. Mar Biotechnol 23:964–975
Shen H, Zhang X, Qian D, Chen J, Xiong J (2022) Pathobiology of Enterocytozoon hepatopenaei (EHP) in shrimp: diagnosis and interpretation from the gut bacterial community. Aquaculture 554:738169
Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP (2006) Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740
Strobl C, Boulesteix A-L, Zeileis A, Hothorn T (2007) Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinformatics 8:25
Wagg C, Dudenhffer JH, Widmer F, Heijden MA (2018) Linking diversity, synchrony and stability in soil microbial communities. Func Ecol 32:1280–1292
Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, Spear J, Caporaso G, Ran B, Knight R (2017) BugBase predicts organism-level microbiome phenotypes. BioRxiv 133462
Xing G, Lu J, Xuan L, Chen J, Xiong J (2022) Sediment prokaryotic assembly, methane cycling, and ammonia oxidation potentials in response to increasing antibiotic pollution at shrimp aquafarm. J Hazard Mater 343:128885
Xiong J (2018) Progress in the gut microbiota in exploring shrimp disease pathogenesis and incidence. Appl Microbiol Biotechnol 102:7343–7350
Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C (2017) Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol 19:1490–1501
Xiong J, Dai W, Qiu Q, Zhu J, Yang W, Li C (2018) Response of host-bacterial colonization in shrimp to developmental stage, environment and disease. Mol Ecol 27:3686–3699
Xiong J, Xuan L, Yu W, Zhu J, Qiu Q, Chen J (2019) Spatiotemporal successions of shrimp gut microbial colonization: high consistency despite distinct species pool. Environ Microbiol 21:1383–1394
Xun W, Liu Y, Li W, Ren Y, Xiong W, Xu Z, Zhang N, Miao Y, Shen Q, Zhang R (2021) Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 9:35
Yao Z, Yang K, Huang L, Huang X, Zhang D (2018) Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Expr 8:120
Yu W, Wu JH, Zhang J, Yang W, Chen J, Xiong J (2018) A meta-analysis reveals universal gut bacterial signatures for diagnosing the incidence of shrimp disease. FEMS Microbiol Ecol 10:fiy147
Zaneveld JR, Mcminds R, Thurber RV (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2:17121
Zhang W, Zhu Z, Chen J, Qiu Q, Xiong J (2021a) Quantifying the importance of abiotic and biotic factors governing the succession of gut microbiota over shrimp ontogeny. Front Microbiol 12:752750
Zhang X, Li X, Lu J, Qiu Q, Chen J, Xiong J (2021b) Quantifying the importance of external and internal sources to the gut microbiota in juvenile and adult shrimp. Aquaculture 531:735910
Zhao Y, Duan C, Zhang X, Chen H, Ye L (2018) Insights into the gut microbiota of freshwater shrimp and its associations with the surrounding microbiota and environmental factors. J Microbiol Biotechnol 28:946–956
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17
Zhu J, Dai W, Qiu Q, Dong C, Zhang J, Xiong J (2016) Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microb Ecol 72:975–985
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32071549), and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Contributions
JX and JC conceived and designed the research. JL, JM, and QX conducted experiments. JL and JM analyzed data. JX contributed analytical tools. JX and JL wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants performed by any of the authors. The shrimp used in this study are complied with the Animal Care and Ethics Committee Policies and Guidelines of Ningbo University.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, J., Mao, J., Qi, X. et al. The assembly of gut microbiota implicates shrimp acute hepatopancreas necrosis disease progression. Appl Microbiol Biotechnol 107, 7489–7500 (2023). https://doi.org/10.1007/s00253-023-12810-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12810-y