Abstract
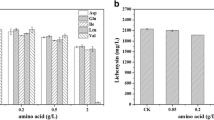
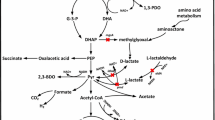
Bacillus licheniformis has been regarded as an outstanding microbial cell factory for the production of biochemicals and enzymes. Due to lack of genetic tools to repress gene expression, metabolic engineering and gene function elucidation are limited in this microbe. In this study, an integrated CRISPR interference (CRISPRi) system was constructed in B. licheniformis. Several endogenous genes, including yvmC, cypX, alsD, pta, ldh, and essential gene rpsC, were severed as the targets to test this CRISPRi system, and the repression efficiencies were ranged from 45.02 to 94.00%. Moreover, the multiple genes were simultaneously repressed with high efficiency using this CRISPRi system. As a case study, the genes involved in by-product synthetic and l-valine degradation pathways were selected as the silence targets to redivert metabolic flux toward l-valine synthesis. Repression of acetolactate decarboxylase (alsD) and leucine dehydrogenase (bcd) led to 90.48% and 80.09 % increases in l-valine titer, respectively. Compared with the control strain DW9i△leuA (1.47 g/L and 1.79 g/L), the l-valine titers of combinatorial strain DW9i△leuA/pHYi-alsD-bcd were increased by 1.27-fold and 2.89-fold, respectively, in flask and bioreactor. Collectively, this work provides a feasible approach for multiplex metabolic engineering and functional genome studies of B. licheniformis.






Similar content being viewed by others
References
Alrumman SA, Mostafa YS, Al-Izran KA, Alfaifi MY, Taha TH, Elbehairi SE (2019) Production and anticancer activity of an l-asparaginase from Bacillus licheniformis isolated from the Red Sea, Saudi Arabia. Sci Rep 9(1):3756. https://doi.org/10.1038/s41598-019-40512-x
Buescher JM, Moco S, Sauer U, Zamboni N (2010) Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem 82(11):4403–4412. https://doi.org/10.1021/ac100101d
Cai D, Rao Y, Zhan Y, Wang Q, Chen S (2019a) Engineering Bacillus for efficient production of heterologous protein: current progress, challenge and prospect. J Appl Microbiol 126(6):1632–1642. https://doi.org/10.1111/jam.14192
Cai D, Zhu J, Zhu S, Lu Y, Zhang B, Lu K, Li J, Ma X, Chen S (2019b) Metabolic engineering of main transcription factors in carbon, nitrogen, and phosphorus metabolisms for enhanced production of bacitracin in Bacillus licheniformis. ACS Synth Biol 8(4):866–875. https://doi.org/10.1021/acssynbio.9b00005
Choi KR, Lee SY (2016) CRISPR technologies for bacterial systems: current achievements and future directions. Biotechnol Adv 34(7):1180–1209. https://doi.org/10.1016/j.biotechadv.2016.08.002
Choudhary E, Thakur P, Pareek M, Agarwal N (2015) Gene silencing by CRISPR interference in mycobacteria. Nat Commun 6:6267. https://doi.org/10.1038/ncomms7267
Cleto S, Jensen JV, Wendisch VF, Lu TK (2016) Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol 5(5):375–385. https://doi.org/10.1021/acssynbio.5b00216
Cress BF, Toparlak OD, Guleria S, Lebovich M, Stieglitz JT, Englaender JA, Jones JA, Linhardt RJ, Koffas MA (2015) CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth Biol 4(9):987–1000. https://doi.org/10.1021/acssynbio.5b00012
Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D (2018) A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat Commun 9(1):1912. https://doi.org/10.1038/s41467-018-04209-5
Diomande SE, Nguyen-The C, Guinebretiere MH, Broussolle V, Brillard J (2015) Role of fatty acids in Bacillus environmental adaptation. Front Microbiol 6:813. https://doi.org/10.3389/fmicb.2015.00813
Ge Y, Li K, Li L, Gao C, Zhang L, Ma C, Xu P (2016) Contracted but effective: production of enantiopure 2, 3-butanediol by thermophilic and GRAS Bacillus licheniformis. Green Chem 18(17):4693–4703. https://doi.org/10.1039/C6GC01023G
Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17(1):148. https://doi.org/10.1186/s13059-016-1012-2
Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H (2012) Improvement of the redox balance increases L-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl Environ Microbiol 78(3):865–875. https://doi.org/10.1128/AEM.07056-11
Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H (2013) Engineering of Corynebacterium glutamicum for high-yield L-valine production under oxygen deprivation conditions. Appl Environ Microbiol 79(4):1250–1257. https://doi.org/10.1128/AEM.02806-12
Jensen ED, Ferreira R, Jakočiūnas T, Arsovska D, Zhang J, Ding L, Smith JD, David F, Nielsen J, Jensen MK (2017) Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb Cell Factories 16(1):46. https://doi.org/10.1186/s12934-017-0664-2
Kim SK, Seong W, Han GH, Lee DH, Lee SG (2017) CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Factories 16(1):188. https://doi.org/10.1186/s12934-017-0802-x
Li X, Wang D, Cai D, Zhan Y, Wang Q, Chen S (2017) Identification and high-level production of pulcherrimin in Bacillus licheniformis DW2. Appl Biochem Biotechnol 183(4):1323–1335. https://doi.org/10.1007/s12010-017-2500-x
Li K, Cai D, Wang Z, He Z, Chen S (2018a) Development of an efficient genome editing tool in Bacillus licheniformis using CRISPR-Cas9 Nickase. Appl Environ Microbiol 84(6). https://doi.org/10.1128/AEM.02608-17
Li Y, Jin K, Zhang L, Ding Z, Gu Z, Shi G (2018b) Development of an inducible secretory expression system in Bacillus licheniformis based on an engineered xylose operon. J Agric Food Chem 66(36):9456–9464. https://doi.org/10.1021/acs.jafc.8b02857
Liang C, Huo Y, Qi G, Wei X, Wang Q, Chen S (2015) Enhancement of L-valine production in Bacillus licheniformis by blocking three branched pathways. Biotechnol Lett 37(6):1243–1248. https://doi.org/10.1007/s10529-015-1783-7
Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang JR, Veening JW (2017) High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13(5):931. https://doi.org/10.15252/msb.20167449
Lv L, Ren YL, Chen JC, Wu Q, Chen GQ (2015) Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P(3HB-co-4HB) biosynthesis. Metab Eng 29:160–168. https://doi.org/10.1016/j.ymben.2015.03.013
Nickel M, Homuth G, Böhnisch C, Mäder U, Schweder T (2004) Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability. Mol Gen Genomics 272(1):98–107
Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA (2016) A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165(6):1493–1506. https://doi.org/10.1016/j.cell.2016.05.003
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183. https://doi.org/10.1016/j.cell.2013.02.022
Qiu Y, Xiao F, Wei X, Wen Z, Chen S (2014) Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol 98(21):8895–8903. https://doi.org/10.1007/s00253-014-5978-y
Qiu Y, Zhang J, Li L, Wen Z, Nomura CT, Wu S, Chen S (2016) Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnology for Biofuels 9:117. https://doi.org/10.1186/s13068-016-0522-1
Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, Fortune SM (2017) Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2:16274. https://doi.org/10.1038/nmicrobiol.2016.274
Takpho N, Watanabe D, Takagi H (2018) High-level production of valine by expression of the feedback inhibition-insensitive acetohydroxyacid synthase in Saccharomyces cerevisiae. Metab Eng 46:60–67. https://doi.org/10.1016/j.ymben.2018.02.011
Tan SZ, Reisch CR, Prather KLJ (2018) A robust CRISPR interference gene repression system in Pseudomonas. J Bacteriol 200(7). https://doi.org/10.1128/JB.00575-17
Tian T, Kang JW, Kang A, Lee TS (2019) Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli. ACS Synth Biol 8(2):391–402. https://doi.org/10.1021/acssynbio.8b00429
Wang D, Wang Q, Qiu Y, Nomura CT, Li J, Chen S (2017) Untangling the transcription regulatory network of the bacitracin synthase operon in Bacillus licheniformis DW2. Res Microbiol 168(6):515–523. https://doi.org/10.1016/j.resmic.2017.02.010
Wang J, Zhao P, Li Y, Xu L, Tian P (2018a) Engineering CRISPR interference system in Klebsiella pneumoniae for attenuating lactic acid synthesis. Microb Cell Factories 17(1):56. https://doi.org/10.1186/s12934-018-0903-1
Wang X, Zhang H, Quinn PJ (2018b) Production of L-valine from metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 102(10):4319–4330. https://doi.org/10.1007/s00253-018-8952-2
Waschkau B, Waldeck J, Wieland S, Eichstadt R, Meinhardt F (2008) Generation of readily transformable Bacillus licheniformis mutants. Appl Microbiol Biotechnol 78(1):181–188. https://doi.org/10.1007/s00253-007-1278-0
Wei X, Ji Z, Chen S (2010) Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Appl Biochem Biotechnol 160(5):1332–1340. https://doi.org/10.1007/s12010-009-8681-1
Westbrook AW, Moo-Young M, Chou CP (2016) Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl Environ Microbiol 82(16):4876–4895
Westbrook AW, Ren X, Moo-Young M, Chou CP (2018) Metabolic engineering of Bacillus subtilis for L-valine overproduction. Biotechnol Bioeng 115(11):2778–2792. https://doi.org/10.1002/bit.26789
Woolston BM, Emerson DF, Currie DH, Stephanopoulos G (2018) Rediverting carbon flux in Clostridium ljungdahlii using CRISPR interference (CRISPRi). Metab Eng 48:243–253. https://doi.org/10.1016/j.ymben.2018.06.006
Wu J, Yu O, Du G, Zhou J, Chen J (2014) Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from l-tyrosine in Escherichia coli. Appl Environ Microbiol 80(23):7283–7292. https://doi.org/10.1128/AEM.02411-14
Wu Y, Chen T, Liu Y, Lv X, Li J, Du G, Ledesma-Amaro R, Liu L (2018) CRISPRi allows optimal temporal control of N-acetylglucosamine bioproduction by a dynamic coordination of glucose and xylose metabolism in Bacillus subtilis. Metab Eng 49:232–241. https://doi.org/10.1016/j.ymben.2018.08.012
Zhan Y, Sheng B, Wang H, Shi J, Cai D, Yi L, Yang S, Wen Z, Ma X, Chen S (2018) Rewiring glycerol metabolism for enhanced production of poly-γ-glutamic acid in Bacillus licheniformis. Biotechnol Biofuels 11(1):306. https://doi.org/10.1186/s13068-018-1311-9
Zhang JL, Peng YZ, Liu D, Liu H, Cao YX, Li BZ, Li C, Yuan YJ (2018) Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb Cell Fact 17(1):62. https://doi.org/10.1186/s12934-018-0909-8
Zhao Y, Li L, Zheng G, Jiang W, Deng Z, Wang Z, Lu Y (2018) CRISPR/dCas9-mediated multiplex gene repression in Streptomyces. Biotechnol J 13(9):e1800121. https://doi.org/10.1002/biot.201800121
Zhou C, Liu H, Yuan F, Chai H, Wang H, Liu F, Li Y, Zhang H, Lu F (2019) Development and application of a CRISPR/Cas9 system for Bacillus licheniformis genome editing. Int J Biol Macromol 122:329–337. https://doi.org/10.1016/j.ijbiomac.2018.10.170
Zuo S, Xiao J, Zhang Y, Zhang X, Nomura CT, Chen S, Wang Q (2018) Rational design and medium optimization for shikimate production in recombinant Bacillus licheniformis strains. Process Biochem 66:19–27. https://doi.org/10.1016/j.procbio.2017.12.012
Funding
This study was funded by the National Key Research and Development Program of China (No. 2018YFA0900303), the National Natural Science Foundation of China (31972849), the Technical Innovation Special Fund of Hubei Province (No. 2018ACA149), and the Natural Science Foundation of Hubei Province of China (2019CFB319).
Author information
Authors and Affiliations
Contributions
S Chen and Y Zhan designed and supervised the study. Y Zhan, P Zheng, M He, and S Sun performed the experiments. Y Zhan and Y Xu analyzed the data and wrote the manuscript. D Wang, D Cai, X Ma, and S Chen revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 664 kb)
Rights and permissions
About this article
Cite this article
Zhan, Y., Xu, Y., Zheng, P. et al. Establishment and application of multiplexed CRISPR interference system in Bacillus licheniformis. Appl Microbiol Biotechnol 104, 391–403 (2020). https://doi.org/10.1007/s00253-019-10230-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10230-5