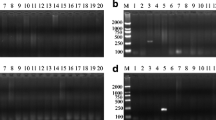
Abstract
The identification of animal species in feed and feedstuffs is important for detecting contamination and fraudulent replacement of animal components that might cause health and economic problems. A novel multiplex assay, based on xMAP technology and the generic detection of closely related species, was developed for the simultaneous differential detection of avian, fish, and ruminant DNA in products. Universal primers and probes specific to avian, fish, or ruminant species were designed to target a conserved mitochondrial DNA sequence in the 12S ribosomal RNA gene (rRNA). The assay specificity was validated using samples of 27 target and 10 nontarget animal species. The limits of detection of the purified DNA were determined to be 0.2 pg/μL–0.1 ng/μL by testing the meat samples of six species and four feedstuffs. The detection sensitivity of the experimental mixtures was demonstrated to be 0.01% (weight percentage). The assay’s suitability for practical application was evaluated by testing feed samples; unlabeled animal ingredients were detected in 32% of the 56 samples. The assay differentially detected the three targeted categories of animal species in less than 2 h, reflecting improvements in speed and efficiency. Based on these results, this novel multiplex xMAP assay provides a reliable and highly efficient technology for the routine detection of animal species in feed and other products for which this information is needed.

Similar content being viewed by others
References
Aristoy MC, Toldra F (2004) Histidine dipeptide HPLC-based test for the detection of mammalian origin proteins in feeds for ruminants. Meat Sci 67(2):211–217
Babady NE, Miranda E, Gilhuley KA (2011) Evaluation of Luminex xTAG fungal analyte-specific reagents for rapid identification of clinically relevant fungi. J Clin Microbiol 49:3777–3782
Barakat H, El-Garhy HA, Moustafa MM (2014) Detection of pork adulteration in processed meat by species-specific PCR-QIAxcel procedure based on D-loop and cytb genes. Appl Microbiol Biotechnol 98(23):9805–9816
Bertolini F, Ghionda MC, D’Alessandro E, Geraci C, Chiofalo V, Fontanesi L (2015) A next generation semiconductor based sequencing approach for the identification of meat species in DNA mixtures. PLoS One 10(4):e0121701. https://doi.org/10.1371/journal.pone.0121701
Cawthraw S, Saunders GC, Martin TC, Sawyer J, Windl O, Reaney SD (2009) Real-time PCR detection and identification of prohibited mammalian and avian material in animal feed. J Food Prot 72(5):1055–1062
Chen R, Yu X-L, Gao X-B, Xue C-Y, Song C-X, Li Y, Cao Y-C (2015) Bead-based suspension array for simultaneous differential detection of five major swine viruses. Appl Microbiol Biotechnol 99:919–928
Cho AR, Dong HJ, Cho S (2014) Meat species identification using loop-mediated isothermal amplification assay targeting species-specific mitochondrial DNA. Korean J Food Sci An 34(6):799–807
Dalmasso A, Fontanella E, Piatti P, Civera T, Rosati S, Bottero MT (2004) A multiplex PCR assay for the identification of animal species in feedstuffs. Mol Cell Probes 18:81–87
Deb R, Sengar GS, Singh U, Kumar S, Alyethodi RR, Alex R, Raja TV, Das AK, Prakash B (2016) Application of a loop-mediated isothermal amplification assay for rapid detection of cow components adulterated in buffalo milk/meat. Mol Biotechnol 58:850–860
Dooley JJ, Paine KE, Garrett SD, Brown HM (2004) Detection of meat species using TaqMan real-time PCR assays. Meat Sci 68:431–438
Dunbar SA (2006) Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta 363:71–82
Dunbar SA, Ritchie VB, Hoffmeyer MR, Rana GS, Zhang H (2015) Luminex® multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods Mol Biol 1225:1–27
European Commission (EC) (1998) Commission Decision 98/88/EC of 13 November 1998 establishing guideline for microscopic identification and estimation of constituents of animal origin for the official control of feeding stuffs. Off J Eur Communities L 318:45–50
Floren C, Wiedemann I, Brenig B, Schütz E, Beck J (2015) Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem 173:1054–1058
Giaretta N, Di Giuseppe AM, Lippert M, Parente A, Di Maro A (2013) Myoglobin as marker in meat adulteration: a UPLC method for determining the presence of pork meat in raw beef burger. Food Chem 141(3):1814–1820
Hamza IA, Jurzik L, Wilhelm M (2014) Development of a luminex assay for the simultaneous detection of human enteric viruses in sewage and river water. J Virol Methods 204:65–72
Kim SH, Huang TS, Seymour TA, Wei CI, Kempf SC, Bridgman CR, Momcilovic D, Clemens RA, An H (2005) Development of immunoassay for detection of meat and bone meal in animal feed. J Food Prot 68(9):1860–1865
Koh BRD, Kim JY, Na HM, Park SD, Kim YH (2011) Development of species-specific multiplex PCR assays of mitochondrial 12S rRNA and 16S rRNA for the identification of animal species. Korean J Vet Serv 34:417–428
Krcmar P, Rencova E (2005) Quantitative detection of species-specific DNA in feedstuffs and fish meals. J Food Prot 68(6):1217–1221
Lin CC, Fung LL, Chan PK, Lee CM, Chow KF, Cheng SH (2014) A rapid low-cost high-density DNAbased multi-detection test for routine inspection of meat species. Meat Sci 96(2):922–929
Maine IR, Atterbury R, Chang KC (2015) Investigation into the animal species contents of popular wet pet foods. Acta Vet Scand 57:7. https://doi.org/10.1186/s13028-015-0097-z
Mandrile L, Amato G, Marchis D, Martra G, Rossi AM (2017) Species-specific detection of processed animal proteins in feed by Raman spectroscopy. Food Chem 229:268–275
OIE (2018) The control of hazards of animal health and public health importance in animal feed. In: Terrestrial Animal Health Code 2018, Chapter 6.4. http://www.oie.int/en/standard-setting/terrestrial-code/access-online/ Accessed 11 December 2018
Pafundo S, Busconi M, Agrimonti C, Fogher C, Marmiroli N (2010) Storage-time effects on olive oil DNA assessed by amplified fragments length polymorphisms. Food Chem 123(3):787–793
Ponzoni E, Breviario D, Mautino A, Giani S, Morello L (2013) A multiplex, bead-based assay for profiling plant-derived components in complex food matrixes. Anal Bioanal Chem 405:9849–9858
Rao Q, Hsieh YH (2008) Competitive enzyme-linked immunosorbent assay for quantitative detection of bovine blood in heat-processed meat and feed. J Food Prot 71(5):1000–1006
Reslova N, Michna V, Kasny M, Mikel P, Kralik P (2017) xMAP technology: applications in detection of pathogens. Front Microbiol 8:55. https://doi.org/10.3389/fmicb.2017.00055
Rodríguez MA, García T, González I, Asensio L, Hernández PE, Martín R (2004) Quantitation of mule duck in goose foie gras using TaqMan real-time polymerase chain reaction. J Agric Food Chem 52(6):1478–1483
Ros-García A, Barandika JF, García-Pérez AL, Juste RA, Hurtado A (2013) Assessment of exposure to piroplasms in sheep grazing in communal mountain pastures by using a multiplex DNA bead-based suspension array. Parasit Vectors 6:277. https://doi.org/10.1186/1756-3305-6-277
Shepherd CJ, Jackson AJ (2013) Global fishmeal and fish-oil supply: inputs, outputs and markets. J Fish Biol 83(4):1046–1066. https://doi.org/10.1111/jfb.12224
World Health Organization (WHO) (1996) Report on a WHO consultation on public health issues related to human and animal transmissible spongiform encephalopathies (with the participation of Food and Agriculture Organization and Office International des Epizooties). www.who.int/csr/disease/bse/variantCJD/en/ Accessed 30 January 2003
Wu Y, Yang Y, Liu M, Wang B, Han J, Chen Y (2016) A 15-plex/xMAP methods to detect 15 animal ingredients by suspension array system coupled with multifluorescent magnetic beads. J AOAC Int 99(3):1–10
Acknowledgments
We thank Hai-Yan Luo and Xin Tan for technical assistance.
Funding
This work was supported by the Science Research Program of Guangzhou, China (grant no. 201707010487) and the Science and Technology Research Project of the former Guangdong Entry-exit Inspection and Quarantine Bureau, China (grant no. 2018GDK04).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, R., Gao, XB., Mei, MZ. et al. A novel multiplex xMAP assay for generic detection of avian, fish, and ruminant DNA in feed and feedstuffs. Appl Microbiol Biotechnol 103, 4575–4584 (2019). https://doi.org/10.1007/s00253-019-09833-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09833-9