Abstract
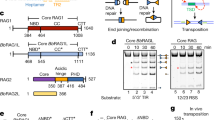
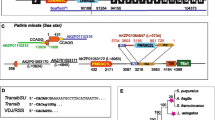
RAG1 and RAG2 are essential subunits of the V(D)J recombinase required for the generation of the variability of antibodies and T cell receptors in jawed vertebrates. It was demonstrated that the amphioxus homologue of RAG1-RAG2 is encoded in an active transposon, belonging to the transposase DDE superfamily. The data provided support the possibility that the RAG transposon has been active through the deuterostome evolution and is still active in several lineages. The RAG transposon corresponds to several families present in deuterostomes. RAG1-RAG2 V(D)J recombinase evolved from one of them, partially due to the new ability of the transposon to interact with the cellular reparation machinery. Considering the fact that the RAG transposon survived millions of years in many different lineages, in multiple copies, and that DDE transposases evolved their association with proteins involved in repair mechanisms, we propose that the apparition of V(D)J recombination machinery could be a predictable genetic event.





Similar content being viewed by others
References
Agrawal A et al (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744–751
Altschul SF et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Chatterji M et al (2006) Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol 26:1558–1568
Curry JD et al (2007) Chromosomal reinsertion of broken RSS ends during T cell development. J Exp Med 204:2293–2303
Danchin E et al (2004) The major histocompatibility complex origin. Immunol Rev 198:216–232
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331–368
Fugmann SD (2010) The origins of the Rag genes from transposition to V(D)J recombination. Semin Immunol 22:10–16
Fugmann SD et al (2006) An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A 103:3728–3733
Hiom K et al (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463–470
Hsu E, Lewis SM (2015) The origin of V(D)J diversification. In: Alt FW, Honjo T, Radbruch A, Reth M (eds) Molecular biology of B cells. Elsevier, Amsterdam, pp 133–148
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huang S et al (2016) Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166:102–114
Izsvák Z et al (2004) Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell 13:279–290
Kapitonov VV, Jurka J (2005) RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3:998–1011
Kapitonov VV, Jurka J (2007) Chapaev—a novel superfamily of DNA transposons. Repbase Reports 7:777–777
Kapitonov V.V., Koonin E.V. (2015) Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol. Direct, http://biologydirect.biomedcentral.com/articles/10.1186/s13062-015-0055-8.
Kim MS et al (2015) Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 518:507–511
Koonin EV, Krupovic M (2015) Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet 16:184–192
Panchin Y, Moroz LL (2008) Molluscan mobile elements similar to the vertebrate recombination-activating genes. Biochem Biophys Res Commun 369:818–823
Ramsden DA, Weed BD, Reddy YV et al (2010) V(D)J recombination: born to be wild. Semin Cancer Biol 20:254–260
Ru H et al (2015) Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell 163:1138–1152
Solovyev V et al (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7:S10.1–S10.12
Tamura K et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Teng G, Schatz DG (2015) Regulation and evolution of the RAG recombinase. Adv Immunol 128:1–39
Vanura K. et al. (2007) In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability. PLoS Biol. 5, http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050043
Warbrick E et al (1998) PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res 26:3925–3932
Acknowledgments
We thank the EBM laboratory for advice and Olivier Loison and Ruth Heyns for editing the manuscript and the help provided with the articles images.
Author information
Authors and Affiliations
Contributions
JRMP, PP and SFH conceived the project and design the study. JRMP, PP and SFH analyzed the results. JRMP, PP, SFH and ALX wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Figure S1
Features of the proteins encoded by the RAG and RAG-like proteins. (A) Protein alignment of RAG1L with vertebrate RAG1. Repeat motifs in amphioxus and the purple sea urchin RAG1L were removed and replaced with an “X” and highlighted in yellow. Three regions of conserved cysteine and histidine residues that might bind zinc are underlined with green bars. The N-terminal zinc binding dimerization domain is underlined with dark-red bars. The subdomains of the RAG1 core region are indicated with colored bars. The conserved acidic catalytic residues are highlighted with red shading (D600, E662, D708 and E962 on mouse RAG1). The PflRAG1LA is more similar to vertebrate RAG1, and those regions were labeled with “*”. GenBank accessions for mouse RAG1, shark RAG1, lancelet RAG2L and sea urchin RAG1L are NP_033045, XP_007886047, KJ748699 and NP_001028179, respectively. (B) Protein alignment of RAG2L with vertebrate RAG2. Color shading shows the conservation of physiochemical properties. The N-terminal amino acid sequences correspond to Kelch-like repeats. The central conserved GG motifs of the six Kelch-like repeats are underlined in red. The plant homeodomain (PHD) is also underlined below the alignment. GenBank accessions for mouse RAG2, shark RAG2, lancelet RAG2L and sea urchin RAG2L are NP_033046, XP_007885835, KJ748699 and NP_001028184, respectively. (PDF 393 kb)
Figure S2
Percent Identity Matrix of RAG1 (S2A) and RAG2 (S2B). In order to provide a multiple alignment, Clustal-Omega requires a guide tree which defines the order in which sequences/profiles are aligned. A guide tree in turn is constructed, based on a distance matrix. Conventionally, this distance matrix is comprised of all the pairwise distances of the sequences. The distance measure Clustal-Omega uses for pairwise distances of unaligned sequences is the k-tuple measure. By default, the distance matrix is used internally to construct the guide tree and is then discarded. By specifying, the internal distance matrix can be written to file. (PDF 210 kb)
Table S1
RAGL distribution in non-chordate genome and expressed sequence. Distribution in the cephalordate phyla: B. belcheri and B. floridae available in 13. (PDF 230 kb)
Rights and permissions
About this article
Cite this article
Morales Poole, J.R., Huang, S.F., Xu, A. et al. The RAG transposon is active through the deuterostome evolution and domesticated in jawed vertebrates. Immunogenetics 69, 391–400 (2017). https://doi.org/10.1007/s00251-017-0979-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-0979-5