Abstract
Circulating cell-free DNA (ccfDNA) has great potential for non-invasive diagnostics, and prediction and monitoring of treatment response, but its amount is usually limited. Therefore, the choice of methods to extract and characterize ccfDNA is crucial. In the current study, we performed the most comprehensive comparison of methods for ccfDNA extraction (11 methods), quantification (3 methods), and estimation of the integrity index (2 methods) from small quantities of different kinds of plasma. The QIAamp® Circulating Nucleic Acid Kit and the Norgen Plasma/Serum Circulating DNA Purification Mini Kit showed the best accuracy and reproducibility, but the Norgen kit allowed to extract a higher amount of ccfDNA. This workflow provides a reliable protocol for the multiple applications of ccfDNA in biomedicine.

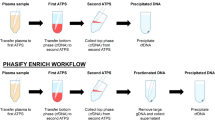
Workflow for the evaluation of methods to isolate, quantify and characterize circulating cell-free DNA from small volumes of plasma


Similar content being viewed by others
References
Esposito A, Bardelli A, Criscitiello C, Colombo N, Gelao L, Fumagalli L, Minchella I, Locatelli M, Goldhirsch A, Curigliano G (2014) Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev 40:648–655
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497:108–112
Chan RW, Jiang P, Peng X, Tam LS, Liao GJ, Li EK, Wong PC, Sun H, Chan KC, Chiu RW, Lo YM (2014) Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc Natl Acad Sci U S A 111:E5302–E5311
Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, Huggett JF (2014) Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 406:6499–6512
Jung K, Fleischhacker M, Rabien A (2010) Cell-free DNA in the blood as a solid tumor biomarker—a critical appraisal of the literature. Clin Chim Acta 411:1611–1624
Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 1775:181–232
Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV (2013) The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci 14:18925–18958
Fong SL, Zhang JT, Lim CK, Eu KW, Liu Y (2009) Comparison of 7 methods for extracting cell-free DNA from serum samples of colorectal cancer patients. Clin Chem 55:587–589
Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, Blighe K, Marchese SD, Hills A, Woodley L, Stebbing J, Coombes RC, Shaw JA (2013) Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 8:e77963
Pinzani P, Salvianti F, Zaccara S, Massi D, De Giorgi V, Pazzagli M, Orlando C (2011) Circulating cell-free DNA in plasma of melanoma patients: qualitative and quantitative considerations. Clin Chim Acta 412:2141–2145
Salvianti F, Pinzani P, Verderio P, Ciniselli CM, Massi D, De Giorgi V, Grazzini M, Pazzagli M, Orlando C (2012) Multiparametric analysis of cell-free DNA in melanoma patients. PLoS One 7:e49843
Zaher ER, Anwar MM, Kohail HMA, El-Zoghby SM, Abo-El-Eneen MS (2012) Value of circulating DNA concentration and integrity as a screening test for detection of cancer in an Egyptian cohort. Alex J Med 48:187–196
Schmidt B, Weickmann S, Witt C, Fleischhacker M (2005) Improved method for isolating cell-free DNA. Clin Chem 51:1561–1563
Yuan H, Zhu ZZ, Lu Y, Liu F, Zhang W, Huang G, Zhu G, Jiang B (2012) A modified extraction method of circulating free DNA for epidermal growth factor receptor mutation analysis. Yonsei Med J 53:132–137
Hufnagl C, Stöcher M, Moik M, Geisberger R, Greil R (2013) A modified phenol-chloroform extraction method for isolating circulating cell free DNA of tumor patients. J Nucleic Acids Investig 4:1
Xue X, Teare MD, Holen I, Zhu YM, Woll PJ (2009) Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta 404:100–104
Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B, Del Rio M, Lamy PJ, Bibeau F, Nouaille M, Loriot V, Jarrousse AS, Molina F, Mathonnet M, Pezet D, Ychou M (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20:430–435
Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, Rio Frio T, Pierron G, Callens C, Bieche I, Saliou A, Madic J, Rouleau E, Bidard FC, Lantz O, Stern MH, Le Tourneau C, Pierga JY (2015) Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 9:783–90
How-Kit A, Lebbe C, Bousard A, Daunay A, Mazaleyrat N, Daviaud C, Mourah S, Tost J (2014) Ultrasensitive detection and identification of BRAF V600 mutations in fresh frozen, FFPE, and plasma samples of melanoma patients by E-ice-COLD-PCR. Anal Bioanal Chem 406:5513–5520
Acknowledgments
The authors would like to thank Chao Feng (CNG) for his help with the statistical analysis and Steven McGinn (CNG) for the critical reading of the manuscript. This work was partly supported by the EU FP7 under grant agreement number 241669 (the CAGEKID project, http://www.cng.fr/cagekid).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 237 kb)
Rights and permissions
About this article
Cite this article
Mauger, F., Dulary, C., Daviaud, C. et al. Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem 407, 6873–6878 (2015). https://doi.org/10.1007/s00216-015-8846-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-8846-4