Abstract
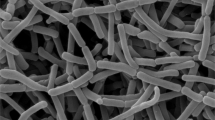
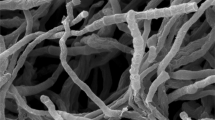
A novel actinomycete, strain PA1-10T, isolated from the leaf of Phyllanthus amarus collected from Bangkok, Thailand, was characterized taxonomically using a polyphasic approach. This strain contained the characteristics consistent with those of members of the genus Nonomuraea. It formed short rugose spore chain on aerial mycelium. The diamino acid in cell wall peptidoglycan was meso-diaminopimelic acid. Galactose, glucose, madurose, mannose, and ribose were found in whole-cell hydrolysates. Predominant menaquinones were MK-9 (H2), MK-9 (H4), and MK-9 (H6). Major cellular fatty acids were iso-C16:0 and C17:0 10-methyl. Phospholipid profiles were composed of phosphatidylinositol mannoside (PIM), lyso-phosphatidylethanolamine (lyso-PE), phosphatidylethanolamine (PE), methylphosphatidylethanolamine (PME), diphosphatidylglycerol (DPG), and phosphatidylglycerol (PG). The G + C content of DNA was 71.2 mol%. Strain PA1-10T showed the highest 16S rRNA gene sequence similarity with Nonomuraea candida JCM 15928T (98.35%) and shared the same node with Nonomuraea maritima JCM 18321T in the phylogenetic tree analysis. Based on the phenotypic characteristics, DNA–DNA relatedness, and average nucleotide identity (ANI), the strain is considered to represent a novel species of the genus Nonomuraea, for which the name Nonomuraea phyllanthi is proposed. The type strain is PA1-10T (= JCM 33073T = NBRC 112774T = TISTR 2497T).


Similar content being viewed by others
References
Ara I, Kudo T, Matsumoto A, Takahashi Y, Omura S (2007) Nonomuraea maheshkhaliensis sp. nov., a novel actinomycete isolated from mangrove rhizosphere mud. J Gen Appl Microbiol 53:159–166
Arai T (1975) Culture media for actinomycetes. The Society for Actinomycetes, Tokyo
Aziz RK, Bartels D, Best AA, Dejongh M, Disz T et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS et al (2012) SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Camas M, Sazak A, Spröer C, Klenk HP, Cetin D, Guven K, Sahin N (2013) Nonomuraea jabiensis sp. nov., isolated from arid soil. Int J Syst Evol Microbiol 63(1):212–218
Chiba S, Suzuki M, Ando K (1999) Taxonomic re-evaluation of ‘Nocardiopsis’ sp. K-252T (= NRRL 15532T): a proposal to transfer this strain to the genus Nonomuraea as Nonomuraea longicatena sp. nov. Int J Syst Bacteriol 49:1623–1630
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Huang H, Liu M, Zhong W, Mo K, Zhu J et al (2018) Nonomuraea mangrovi sp. nov., an actinomycete isolated from mangrove soil. Int J Syst Evol Microbiol 68:3144–3148
Kämpfer P (2012) Genus VI. Nonomuraea corrig. Zhang, Wang and Ruan 1998b, 149VP. In: Goodfellow M, Kampfer P, Busse H-J, Trujillo ME, Suzuki K et al (eds) Bergey’s manual of systematic bacteriology, part B, vol 5, 2nd edn. Springer, New York, pp 1844–1861
Kämpfer P, Kroppenstedt RM, Grün-Wollny I (2005) Nonomuraea kuesteri sp. nov. Int J Syst Evol Microbiol 55(2):847–851
Kelly KL (1964) Inter-society color council: National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. US Government Printing Office, Washington
Klykleung N, Tanasupawat S, Pittayakhajonwut P, Ohkuma M, Kudo T (2015) Amycolatopsis stemonae sp. nov., isolated from a Thai medicinal plant. Int J Syst Evol Microbiol 65:3894–3899
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–148
Le Roes M, Meyers PR (2008) Nonomuraea candida sp. nov., a new species from South African soil. Antonie van Leeuwenhoek 3(1–2):133–139
Meier-Kolthoff PJ, Alexander FA, Klenk HP, Göker M (2013) Genome sequence based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Mikami H, Ishida Y (1983) Post-column fluorometric detection of reducing sugar in high-performance liquid chromatography using arginine. Bunseki Kagaku 32:E207–E210
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Niemhom N, Chutrakul C, Suriyachadkun C, Thawai C (2017) Nonomuraea stahlianthi sp. nov., an endophytic actinomycete isolated from the stem of Stahlianthus campanulatus. Int J Syst Evol Microbiol 67:2879–2884
Nonomura H, Ohara Y (1971) Distribution of actinomycetes in soil. XI. Some new species of the genus Actinomadura. J Ferment Technol 49:904–912
Quadri SR, Tian XP, Zhang J, Li J, Nie GX et al (2015) Nonomuraea indica sp. nov., novel actinomycetes isolated from lime-stone open pit mine, India. J Antibiot 68:491–495
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) J SpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI Inc., Newark, p 1990
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16(6):313–340
Sripreechasak P, Phongsopitanun W, Supong K, Pittayakhajonwut P, Kudo T, Ohkuma M, Tanasupawat S (2017) Nonomuraea rhodomycinica sp. nov., isolated from peat swamp forest soil. Int J Syst Evol Microbiol 67:1683–1687
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Sungthong R, Nakaew N (2015) The genus Nonomuraea: a review of a rare actinomycete taxon for novel metabolites. J Basic Microbiol 55:554–565
Tamaoka J (1994) Determination of DNA base composition. In: Goodfellow M, O’Donnell AG (eds) Chemical methods in prokaryotic systematics. Wiley, Chichester, pp 463–470
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang F, Shi J, Huang Y, Wu Y, Deng X (2017) Nonomuraea ceibae sp. nov., an actinobacterium isolated from Ceiba speciosa rhizosphere. Int J Syst Evol Microbiol 67:1158–1162
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O et al (1987) International committee on Systematic Bacteriology. Report of the ad hoc committee on the reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 37:463–464
Williams ST, Cross T (1971) Chapter XI actinomycetes. Methods Microbiol 4:295–334
Xi L, Zhang L, Ruan J, Huang Y (2011) Nonomuraea maritima sp. nov., isolated from coastal sediment. Int J Syst Evol Microbiol 61:2740–2744
Yoon SH, Ha SM, Kown S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequence and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang Z, Wang Y, Ruan J (1998) Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol 48:411–422
Acknowledgements
We would like to thank the Associate Professor Thatree Phadungcharoen, the Faculty of Pharmaceutical Sciences, Chulalongkorn University for the plant sample and the Pharmaceutical Research Instrument Center, Faculty of Pharmaceutical Sciences, Chulalongkorn University, for providing research facilities.
Funding
This study was supported by the Thailand Research Fund for a 2017 Royal Golden Jubilee Ph.D. Program as a scholarship to N. K. and the Grant for International Research Integration, Research Pyramid, Ratchadaphiseksomphot Endowment Fund (GCURP_58_01_33_01), Chulalongkorn University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Klykleung, N., Yuki, M., Kudo, T. et al. Nonomuraea phyllanthi sp. nov., an endophytic actinomycete isolated from the leaf of Phyllanthus amarus. Arch Microbiol 202, 55–61 (2020). https://doi.org/10.1007/s00203-019-01717-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01717-w