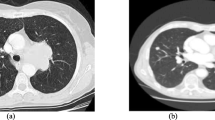
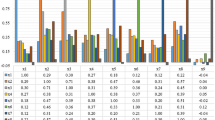
Abstract
Brain tumor classification and segmentation for different weighted MRIs are among the most tedious tasks for many researchers due to the high variability of tumor tissues based on texture, structure, and position. Our study is divided into two stages: supervised machine learning-based tumor classification and image processing-based region of tumor extraction. For this job, seven methods have been used for texture feature generation. We have experimented with various state-of-the-art supervised machine learning classification algorithms such as support vector machines (SVMs), K-nearest neighbors (KNNs), binary decision trees (BDTs), random forest (RF), and ensemble methods. Then considering texture features into account, we have tried for fuzzy C-means (FCM), K-means, and hybrid image segmentation algorithms for our study. The experimental results achieved a classification accuracy of 94.25%, 87.88%, 89.57%, 96.99%, and 97% with SVM, KNN, BDT, RF, and Ensemble methods, respectively, on FLAIR-, T1C-, and T2-weighted MRI, and the hybrid segmentation attaining 90.16% mean dice score for tumor area segmentation against ground-truth images.














Similar content being viewed by others
Availability of Data and Material
Yes.
References
Tandel, Gopal S., Balestrieri, Antonella., Jujaray, Tanay., Khanna, Narender N., Saba, Luca.,Suri, Jasjit S.: Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Computers in Biology and Medicine, 122:103804, (2020)
Padma Nanthagopal, A., Sukanesh Rajamony, R.: Classification of benign and malignant brain tumor ct images using wavelet texture parameters and neural network classifier. J. Vis. 16(1), 19–28 (2013)
Muhammad, S., Salman, K., Khan, M., Wanqing, W., Amin, U., Sung Wook, B.: Multi-grade brain tumor classification using deep cnn with extensive data augmentation. Journal of computational science 30, 174–182 (2019)
Suneetha, B., JhansiRani, A.: A survey on image processing techniques for brain tumor detection using magnetic resonance imaging. In: 2017 International Conference on Innovations in Green Energy and Healthcare Technologies (IGEHT) , pp. 1–6. IEEE (2017)
Pei, L., Bakas, S., Vossough, A., Reza, S.M.S., Davatzikos, C., Iftekharuddin, K.M.: Longitudinal brain tumor segmentation prediction in mri using feature and label fusion. Biomedical Signal Processing and Control 55, 101648 (2020)
Scarapicchia, V., Brown, C., Mayo, C., Gawryluk, J.R.: Functional magnetic resonance imaging and functional near-infrared spectroscopy: insights from combined recording studies. Frontiers in human neuroscience 11, 419 (2017)
Nikolaos P Asimakis, Irene S Karanasiou, PK Gkonis, and Nikolaos K Uzunoglu. Theoretical analysis of a passive acoustic brain monitoring system. Progress in Electromagnetics Research, 23:165–180, 2010
Chaturvedi, C.M., Singh, V.P., Singh, P., Basu, P., Singaravel, M., Shukla, R.K., Dhawan, A., Pati, A.K., Gangwar, R.K., Singh, S.: 2.45 ghz (cw) microwave irradiation alters circadian organization, spatial memory, dna structure in the brain cells and blood cell counts of male mice, mus musculus. Progr. Electromagn. Res. B 29, 23–42 (2011)
Lemieux, L., Hagemann, G., Krakow, K., Woermann, F.G.: Fast, accurate, and reproducible automatic segmentation of the brain in t1-weighted volume mri data. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 42(1), 127–135 (1999)
Tang, H., Wu, E.X., Ma, Q.Y., Gallagher, D., Perera, G.M., Zhuang, T.: MRI brain image segmentation by multi-resolution edge detection and region selection. Comput. Med. Imaging Gr. 24(6), 349–357 (2000)
Chen, V., Ruan, S.: Graph cut based segmentation of brain tumor from mri images. International Journal on Sciences and Techniques of Automatic control & computer engineering 3(2), 1054–1063 (2009)
Jara, H., Sakai, O., Mankal, P., Irving, R.P., Norbash, A.M.: Multispectral quantitative magnetic resonance imaging of brain iron stores: a theoretical perspective. Topics in Magnetic Resonance Imaging 17(1), 19–30 (2006)
Kabir, Y., Dojat, M., Scherrer, B., Forbes, F., Garbay, C.: Multimodal MRI segmentation of ischemic stroke lesions. In: 2007 29th annual international conference of the IEEE engineering in medicine and biology society, pp. 1595–1598 (2007)
Mishra, S.K., Deepthi, VH.: Brain image classification by the combination of different wavelet transforms and support vector machine classification. J. Am. Intell. Human Comput. 12(6), 6741–6749 (2021)
Gumaei, A., Hassan, M.M., Rafiul Hassan, Md., Alelaiwi, A.: Ahybrid feature extractionmethod with regularized extreme learningmachine for brain tumor classification. IEEE Access 7, 36266–36273 (2019)
Mishra, Sonali, Majhi, Banshidhar, Sa, Pankaj Kumar: Texture feature based classification on microscopic blood smear for acute lymphoblastic leukemia detection. Biomed. Signal Process. Control 47, 303–311 (2019)
Amin, J., Sharif, M., Yasmin, M., Fernandes, S.L.: A distinctive approach in brain tumor detection and classification using mri. Pattern Recognition Letters 139, 118–127 (2020)
Bahadure, N.B., Ray, A.K., Thethi, H.P.: Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. International journal of biomedical imaging, 2017 (2017)
Joseph, R.P., Singh, C.S., Manikandan, M.: Brain tumor mri image segmentation and detection in image processing. International Journal of Research in Engineering and Technology 3(1), 1–5 (2014)
Marco, A., Salem, A.B.M.: An automatic classification of brain tumors through MRI using support vector machine. Egy. Comp. Sci. J., 40(3), (2016)
Ahmadvand, A., Kabiri, P.: Multispectral MRI image segmentation using Markov random field model. Signal Image Video Process 10(2), 251–258 (2016)
Islam, A., Reza, S.M.S., Iftekharuddin, K.M.: Multifractal texture estimation for detection and segmentation of brain tumors. IEEE transactions on biomedical engineering 60(11), 3204–3215 (2013)
Abbasi, S., Tajeripour, F.: Detection of brain tumor in 3d mri images using local binary patterns and histogram orientation gradient. Neurocomputing 219, 526–535 (2017)
Işın, A., Direkoğlu, C., Şah, M.: Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Computer Science 102, 317–324 (2016)
Arı, B., Şengür, A., Arı, A.: Local receptive fields extreme learning machine for apricot leaf recognition. In: International Conference on Artificial Intelligence and Data Processing (IDAP16), pp. 17–18 (2016)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, et al, R.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., Freymann, J.B., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Scientific data 4(1), 1–13 (2017)
Spyridon, B., Mauricio, R., Andras, J., Stefan, B., Markus, R., Alessandro, C., Russell, T.S., Christoph, B., Sung, M.H., Martin, R., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprintarXiv: 1811.02629 (2018)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., Egan, A., Yushkevich, P.A., Gee, J.C.: N4itk: improved n3 bias correction. IEEE transactions on medical imaging 29(6), 1310–1320 (2010)
Humied, I.A., Abou-Chadi, F.E.Z., Rashad, M.Z.: A new combined technique for automatic contrast enhancement of digital images. Egypt. Inf. J. 13(1), 27–37 (2012)
Haralick, R.M., Shanmugam, K., Its Hak, D.I.N.S.T.E.I.N.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. 6, 610–621 (1973)
Qurat-Ul-Ain, G.L., Kazmi, S.B., Jaffar, M.A., Mirza, A.M.: Classification and segmentation of brain tumor using texture analysis. In: Recent advances in artificial intelligence, knowledge engineering and data bases, pp. 147-155 (2010)
Chu, A., Sehgal, C.M., Greenleaf, J.F.: Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 11(6), 415–419 (1990)
Tian, S., Bhattacharya, U., Lu, S., Su, B., Wang, Q., Wei, X., Lu, Y., Tan, C.L.: Multilingual scene character recognition with co-occurrence of histogram of oriented gradients. Pattern Recognition 51, 125–134 (2016)
Prakasa, E.: Texture feature extraction by using local binary pattern. INKOM J. 9(2), 45–48 (2016)
Al-Janobi, A.: Performance evaluation of cross-diagonal texture matrix method of texture analysis. Pattern Recognit. 34(1), 171–180 (2001)
He, D.-C., Wang, L.: Simplified texture spectrum for texture analysis. Journal of Communication and Computer 7(8), 44–53 (2010)
Tandel, Gopal S., Biswas, Mainak, Kakde, Omprakash G., Tiwari, Ashish, Suri, Harman S., Turk, Monica, Laird, John R., Asare, Christopher K., Ankrah, Annabel A., Khanna, et al, N.N.: A review on a deep learning perspective in brain cancer classification. Cancers 11(1), 111 (201+9)
Braun, A.C., Weidner, U., Hinz, S.: Classification in high-dimensional feature spaces-assessment using svm, ivm and rvm with focus on simulated enmap data. IEEE J. Sel. Topi. Appl. Earth Obs. Remote Sens. 5(2), 436–443 (2012)
Tan, S.: An effective refinement strategy for KNN text classifier. Exp. Syst. Appl. 30(2), 290–298 (2006)
Tin Kam Ho: A data complexity analysis of comparative advantages of decision forest constructors. Pattern Anal. Appl. 5(2), 102–112 (2002)
Raf Guns and Ronald Rousseau. Recommending research collaborations using link prediction and random forest classifiers. Scientometrics, 101(2), 1461–1473, 2014
Oza, N.C., Tumer, K.: Classifier ensembles: Select real-world applications. Information fusion 9(1), 4–20 (2008)
Dhanalakshmi, P., Kanimozhi, T.: Automatic segmentation of brain tumor using k-means clustering and its area calculation. International Journal of advanced electrical and Electronics Engineering 2(2), 130–134 (2013)
Kalema, K.A., Bukenya, F., Rose, A.A.: A review and analysis of fuzzy-c means clustering techniques. Int J Sci Eng Res 5(11), 1072–7 (2014)
Raja, K.D.: Segmenting images using hybridization of k-means and fuzzy c-means algorithms. In: Introduction to data science and machine learning, IntechOpen (2019)
Sanjay, S., Suraj, S.: Brain tumor segmentation by texture feature extraction with the parallel implementation of fuzzy c-means using CUDA on GPU. In: 2018 5th international conference on Parallel, Distributed and Grid Computing (PDGC), pp. 580–585. IEEE (2018)
Alam, M. S., Rahman, M. M., Hossain, M. A., Islam, M. K., Ahmed, K. M., Ahmed, K. T., Singh BC, Sipon MM,: Automatic human brain tumor detection in MRI image using template-based k means and improved fuzzy c means clustering algorithm. Big Data Cognit Comput 3(2), 27 (2019)
Funding
This study is not funded by any organization or individual.
Author information
Authors and Affiliations
Contributions
First author is the main contributor and supervised by other authors.
Corresponding author
Ethics declarations
Conflicts of interest
All the authors declare that they have no conflict of interest.
Ethics Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Yes.
Consent for Publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jena, B., Nayak, G.K. & Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Machine Vision and Applications 33, 6 (2022). https://doi.org/10.1007/s00138-021-01262-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00138-021-01262-x