Abstract
Novel oxotriazinoindoles (OTIs) were recently reported as highly efficient and selective aldose reductase inhibitors. Here, a series of novel N(2)-substituted oxotriazinoindoles was developed with the aim to investigate molecular interactions within the aldose reductase (ALR2) inhibitor binding site. About twice increased inhibition efficacy of the most efficient derivative 14 (N(2)-CH2CH2COOH) compared to the unsubstituted lead OTI was obtained, yet at the expense of selectivity relative to anti-target aldehyde reductase (ALR1). To explain the major drop in selectivity, observed also in other N(2)-substituted derivatives, in silico molecular modeling approach revealed the role of extra interactions with the residues of Arg309, Arg312 and Met302 located in the additional C-terminal loop of ALR1 missing in ALR2, which can prevent or enhance binding in ALR1. These key findings will be used for development of the next generation of selective OTI inhibitors.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
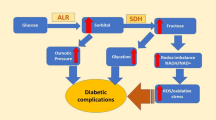
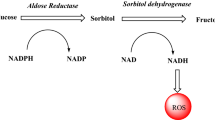
Over the past two decades, despite substantial efforts to unravel the precise physiological role of aldose reductase (AR, ALR2), our comprehension of this enzyme remains limited. AR, as the initial enzyme in the polyol pathway, is recognized as a significant contributor to the development of chronic diabetic complications. In hyperglycaemic conditions AR catalyses the reduction of glucose to sorbitol. The accumulation of sorbitol in cells, coupled with associated metabolic disruptions, leads to complications such as cataracts, peripheral nerve damage, kidney issues, and cardiovascular problems. Compared to aldose reductase (ALR2), aldehyde reductase (ALR1) is considered as detoxification enzyme (an anti-target). These enzymes share approximately 65% sequence identity. Consequently, the search for effective AR inhibitors continues as a promising way for preventing diabetic complications [1,2,3].
Selective inhibition of ALR2 related to ALR1 is crucial due to potential side and unwanted effects associated with several inhibitors of aldose reductase (ARIs). ALR1 catalyses the reduction of highly reactive toxic 2-oxoaldehydes which participate to AGE (Advanced Glycation End products) formation and protein cross-linking. These aldehydes are produced in high amounts under hyperglycaemic conditions, leading to tissue and vascular damage. Simultaneous inhibition of both ALR2 and ALR1 may diminish the benefits of ARIs by preventing the detoxifying function of ALR1. Despite the significant sequence and structural similarities at the active sites of ALR2 and ALR1, there are crucial differences in the substrate-binding site that can be exploited for designing selective inhibitors. Notably, specific amino acid differences in the active sites of ALR2 and ALR1, such as Thr113, Ala299, Leu300 in ALR2 being replaced by Tyr116, Ile299, Pro301 in ALR1, provide potential targets for selectivity. Additionally, an extra loop extending from Pro301 to Arg312 in ALR1, along with non-conserved Arg312 and Asp313 in the active site, play a crucial role in ligand binding for ALR1 [4].
In searching for novel chemotypes of aldose reductase inhibitors (ARIs), cemtirestat (CMTI, 2-(3-thioxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid), was designed as a promising lead structure with high aldose reductase inhibitory efficacy, selectivity and antioxidant activity in a previously studies [1,2,3, 5]. In a follow up study [6], the oxotriazinoindole (bio)isoster of cemtirestat, 2-(3-oxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid (OTI), was synthesized and characterized as an ALR2 inhibitor more efficient than CMTI and moreover endowed with markedly increased selectivity (Fig. 1).
Recently, N(2)-benzyl derivative of OTI was used as a starting scaffold to study a series of substituted derivatives containing varied groups in an ortho position on a benzyl moiety. The aim was to exploit additional interactions within an unoccupied ALR2 pocket surrounded by Trp219, Ala299 and Leu301 (see PDB: 4QX4). The derivative containing a planar and the least solvated -CN functional group was found as the most efficient ALR2 inhibitor of the series [7].
In the present study, an aliphatic series of N(2)-substituted oxotriazinoindoles was synthesized with the aim to further survey potential interactions within the above mentioned ALR2 pocket. About twice increased inhibition efficacy of the most efficient derivative 14 (N(2)-CH2CH2COOH) compared to unsubstituted OTI was obtained yet compromised by a marked drop in selectivity relative to ALR1. In silico molecular modeling approaches were used to explain potentially involved interaction patterns responsible for inhibition efficacy and selectivity.
Results and discussion
Drug design
In the present study, we continued in exploration of an empty ALR2 pocket at close quarters to the N(2) position of a triazine ring of inhibitor cemtirestat (PDB: 4QX4). The above mentioned pocket contains the key amino acid residues Trp219, Ala299, Leu301 and Ser302 [7]. Based on the X-ray structure of CMTI in human aldose reductase (AKR1B1) (PDB: 4QX4), we found a possible enhancement of the interaction energy in N(2)-substitution. In addition, the interaction surface preferred substituents with a lipophilic linker and a hydrophilic end, which allows forming interactions mainly with the residues of Ser302 (illustrated in the Supporting Material: 3.Drug Design, Fig. 40 and 41). We were inspired by our previously published derivatives of OTI-1 and OTI-2 characterized by high ALR2 inhibition efficacy (Fig. 2) [6]. In order to explore ability of ALR2 enzyme to create additional ligand-enzyme interactions within the above mentioned pocket, novel N(2)-substituted derivatives 5, 8, 11 and 14 have been designed and prepared (Scheme 1).
Synthesis of predicted ALR2 inhibitors 5, 8, 11 and 14: a EtOH (abs), H2SO4 9.1 mol eq; reflux, 6 h; b 1. NaH, DMF (abs) 0 °C 30 min, then RT 1 or 2 h for 4, 7 and 10, and K2CO3 DMF (abs), 0 °C, 30 min for 13; 2. R-Cl (3) 1.5 mol eq, RT, 1 h for 4, R-Br (6) 1.5 mol eq, RT, 2 h for 7, R-Br (9) 1.5 mol eq, RT, 1 h for 10, R-Br (12) 1.5 mol eq, 80 °C, 4 h for 13; c NaOH / H2O, 80 °C, 1 h for all final compounds 5, 8, 11 and 14
Proposed derivatives 5, 8, 11 and 14 were docked into the AKR1B1 in conformation complexed with CMTI (from PDB: 4QX4). The oxotriazinoindole OTI (Fig. 3A) provided the key interactions with residues 20, 48, 110–111, 122 and 300. More or less, all new ligands copied the position of this compound (Fig. 3B), whereas compound 14 was found to create five hydrogen bonds (HB) – with Tyr48, Trp111, Cys298, Leu300 and Ser302, which is one HB more than was predicted for the lead structure OTI (Tyr48, His110, Trp111 and Leu300). The complexes with other proposed ligands have had similar numbers of hydrogen bonds (three to five). OTI-2, which also possesses two carboxylic groups as 14, was bound also with five hydrogen bonds, but OTI-2 was more solvated than more lipophilic 14 (the calculated solvation energy was −1345 kJ/mol for OTI-2 and −1157 kJ/mol for 14), what indicates a stronger desolvation penalty and less activity for OTI-2.
A Interactions of OTI in the AKR1B1 binding site. Yellow dashed lines denote H-bonds; green lines are hydrophobic interactions and red line denotes π-π interaction. B Positions of ligands OTI (blue), OTI-1 (gray), OTI-2 (green), 11 (yellow), 14 (magenta), 5 (light blue) and 8 (orange) in the binding site of AKR1B1 (taken from PDB: 4QX4). Protein ribbon structure (gray) and visible specified amino acid residues (cyan) are taken from the optimized complex of ALR2 protein with the strongest inhibitor 14
Chemistry
The synthetic ways of predicted compounds 5, 8, 11 and 14 are summarized in Scheme 1.
Esterification of OTI (1) in refluxing dry ethanol under acid catalysis led to the ethyl ester intermediate 2 in 87 % yield. Ester 2 was subsequently N(2)-alkylated to compounds 4, 7, 10 and 13 (73–89 % yield) by reaction of the corresponding alkyl halide under basic conditions. Finally, saponification of the resulting esters 4, 7, 10 and 13 led to the target compounds 5, 8, 11 and 14 in 61, 53, 60 and 54 % overall yields, respectively.
Enzyme inhibition
All synthesized compounds 5, 8, 11 and 14 were tested for their in vitro inhibition activity by reduction of D,L-glyceraldehyde with pre-purified rat lens enzyme ALR2 as well as the efficiency to reduce D-glucuronate by ALR1 enzyme pre-purified from the rat kidneys (Table 1). In the newly synthesized compounds, with exception of 14, the presence of N(2) substituents decreased inhibition efficacy in comparison with the parent OTI that can be explained by their desolvation penalties in case of 14 compensated by additional HB with Ser302 (Fig. 3). For all compounds tested, marked decrease in inhibition selectivity relative to ALR1 was recorded when compared to OTI. In the case of the most efficient ALR2 inhibitor 14, the inhibition efficacy increased about twice, based on the IC50 values, in comparison to OTI, while its selectivity dropped more than 10 times.
Compared to aldose reductase (ALR2), aldehyde reductase (ALR1) is considered as detoxification enzyme (an anti-target). To explain the marked drop in selectivity of the most efficient inhibitor 14 and other derivatives relative to OTI, all compounds were docked also into the binding site of porcine ALR1 (from PDB: 3FX4). As shown in Fig. 4 for compound 14, ALR1 is endowed with one additional loop when compared with aldose reductase. This C-terminal loop, the significance of which was already pointed out by Barski et al. [8, 9] and supported by structure-function studies of Rees Milton et al. [10], Steuber et al. [11] and El-Kabani et al. [12], disposes with the positively charged residues of Arg309 and Arg312. Both arginines can differently anchor the ligands via potential ionic, cation-π and hydrogen bonds. Molecular modeling identified additional interactions of the novel derivatives 5, 8, 11 and 14 substituted in position N(2) which make them less selective than the parent compound OTI (Fig. 4). The blue loop possessing Arg309 of ALR1 (not present in ALR2; orange loop), is visible on both structures shown in Fig. 4 at the bottom left. The border residues of this loop, Arg312 (Ser302 in ALR2) and Met302 (Leu301 in ALR2), create together with Arg309 attractive places for anchoring the studied molecules in ALR1. As shown in Table 1, the substituents in position N(2) can markedly affect inhibition activity and selectivity. The exceptional selectivity of OTI (SF > 2381), shown in its superimposed binding positions on the right picture of Figs. 4 and 5, is given by its anchoring in Arg312 through its only acidic functional group. Strong ionic and cation-π interactions of Arg312 with OTI (blue ligand) in ALR1, keep OTI ligand in higher distance from the active site of ALR1 which causes low inhibition efficacy of OTI (IC50 > 100 000 nM, ALR1). This is in contrast to ALR2 binding of OTI (red ligand) which directly projects its N(5)-CH2COO- group towards an active site of ALR2 possessing also NADP+ cofactor (Fig. 4). A different flipping of OTI in ALR1 and ALR2 explains high observed selectivity by low inhibition of anti-target ALR1 and high inhibition of the target enzyme (ALR2, IC50 = 42 nM). Even though ligand 14 binds in ALR1 via both Arg309 and Arg312, its additional N(2)-CH2CH2COO- group affects its binding position in ALR1 differently (the left picture, blue ligand) when compared to OTI (the right picture, blue ligand). Therefore inhibition of both enzymes ALR1 and ALR2 by 14 is possible, leading to the drop of selectivity factor from > 2 381 for OTI to 231 for 14. As a result, ligand 14 projects its N(5)-CH2COO- group in both ALR1 and ALR2 towards NADP+ cofactor compared to highly selective unsubstituted OTI inhibitor (Fig. 4, two ligands on the right picture). On the other hand, OTI deflects its N(5)-CH2COO- group out of the binding site in ALR1 (low inhibition) and projects it into binding site of ALR2 (high inhibition).
Aligned structures of human AKR1B1 (from PDB: 4QX4; orange ribbon) and porcine ALR1 (from PDB: 3FX4; light blue ribbon) with the positions of ligand 14 (left) and the parent compound OTI (right) in binding site of AKR1B1 (red ligand) and ALR1 (blue ligand). H-bonds are depicted by yellow dashed lines, ionic bond by a magenta line and a cation-π interaction with a green line. The additional loop of ALR1 (blue, denoted by green arrow) with Arg309, Met302 and Arg312 is visible at the bottom
Positions of inactive ligands towards aldehyde reductase OTI (blue), OTI-1 (gray) (A) and the strongest inhibitors of aldehyde reductase OTI-2 (turquoise), 14 (magenta) and 5 (orange) (B) in the binding site of porcine ALR1 (originated from PDB: 3FX4). ALR1 protein ribbon structure (gray) and specified amino acid residues (cyan) visible are taken from the optimized complex of ALR1 with the strongest ALR2 inhibitor 14. Yellow dashed lines denote H-bonds of 14 in ALR1, green thick line is for hydrophobic interaction of ligands with NADP+ skeleton
As indicated in Fig. 5B, the strongest ALR1 inhibitors (OTI-2, 5 and 14) have substituents able to create intermolecular bonds with residues of additional loop Arg309, Arg312 or Met302 in ALR1, but also with those of the catalytic site as His113 and Trp114. On the other hand, the most selective compounds OTI and OTI-1 took similar positions bound firmly with Arg312 (Fig. 5A). As a consequence, they were not able to approach NADP+, while the less selective compounds 5, 14 and OTI-2 interact with NADP+ via hydrophobic and possible π-π and ionic interactions with the coenzyme NADP+ (green thick lines in Fig. 5A).
Predicted positions of all novel compounds in ALR1 were not so unified as in ALR2 (Fig. 6 vs. Fig. 3) which was caused by a lot of potential partners for the carboxymethyl and other novel functional groups at N(2) of the OTI skeleton.
Positions of ligands OTI (blue), OTI-1 (gray), OTI-2 (turquoise), 11 (yellow), 14 (magenta), 5 (orange) and 8 (light blue) in the binding site of porcine ALR1 (originated from PDB: 3FX4, optimized with 14). ALR1 protein ribbon structure (gray) and specified amino acid residues (cyan) visible are taken from the optimized complex of ALR1 with the strongest ALR2 inhibitor 14
The above outcomes are in agreement with our recently published hypothesis [13], that the interaction between a ligand and nicotinamide ring of the cofactor NADP+ is necessary for inhibition of both aldose and aldehyde reductase. Consequently, a potential effect of compounds under screening on releasing of NADP+ and its cyclic reduction should be taken into consideration.
Physicochemical properties and molecular obesity
In the drug design, prediction of pharmacokinetic parameters and molecular obesity is important, which determine the behavior of proposed compounds in the body. Parameters of molecular obesity together with physicochemical properties were calculated for the novel derivatives 5, 8, 11 and 14 in comparison with previously published OTIs, CMTI and epalrestat (Table 2). Calculated values of the ligand efficiency (LE) and the binding efficiency index (BEI) were found in the range of optimal values (> 0.3 and > 14.7, respectively) [14,15,16,17,18]. High inhibition efficacy characterized by pIC50 values close to 7 in combination with low values of logP resulted in the lipophilic LE values (LLE = pIC50 − log P) above the lower limit for successful lead 3.8 [18]. The LLE figures above 6 point to the preference of specific binding to AR. As shown in Table 2, the ligand-efficiency-dependent lipophilicity (LELP) values are in the recommended range < 7.5 [16, 17]. Moreover, N(2) substitution of all tested compound 5, 8, 11 and 14 in comparison with OTI resulted in decrease of the LELP index from 3.4 to around 1. The drop in the LELP index supports the preferred role of specific component in binding of tested compounds to the enzyme. The most active derivative 14 with one additional predicted hydrogen bond in active site of ALR2 was found with LELP value decreased to 0.62 in comparison for the lead structure OTI. All of the compounds shown in Table 2 meet the threshold criteria of the topological polar surface area (TPSA) (60–140 Å2) for good oral absorption [15].
Estimation of passive gastrointestinal absorption
Estimation of passive gastrointestinal absorption was performed using biopartitioning micellar chromatography (BMC), at two pH values (3.0 and 5.5). Biopartitioning micellar chromatography is considered a simple and reliable method for the estimation of passive gastrointestinal absorption, particularly useful in early phases of drug discovery. Higher retention in this chromatographic system indicates better potential of a drug to cross the gastrointestinal lipid bilayer and higher expected passive gastrointestinal absorption [19, 20]. These pH values were selected in order to simulate physiological conditions in stomach and upper small intestine, which are considered the most significant parts of gastrointestinal tract for drug absorption (pH value in stomach is between 1.5 and 3.5, while pH value in upper small intestine is between 5.0 and 6.0) [21,22,23]. To simulate stomach, pH = 3.0 was selected in order to prevent damage of the HPLC column (according to the column manufacturer, recommended mobile phase pH range is 2.0–11.5). The column temperature was set to mimic physiological temperature (36.5 °C). Retention factors of tested compounds are presented in Table 3.
It can be concluded that compounds 5, 8, 11 and 14, as well as standards OTI, OTI-1 and CMTI have significantly lower retention factors than epalrestat at both pH values and their lower passive gastrointestinal absorption can be expected. Furthermore, their retention factors are higher at pH 3.0 than at pH 5.5 and it can be concluded that stomach is the most favorable part of gastrointestinal tract for their passive gastrointestinal absorption. At pH 3.0, ionization of carboxylic group is suppressed (pKa of carboxylic group is 4.14 or 4.30, as presented in Table 2) and dominant forms are unionized ones, which explains higher retention factors and better expected passive gastrointestinal absorption at lower pH values, i.e. in stomach. Taking into account the influence of carboxylic group, its masking (e.g. by esterification or isosteric replacements) could be a valuable strategy to improve their passive gastrointestinal absorption.
Conclusions
The series of N(2)-substituted oxotriazinoindoles was synthesized with the aim to survey in a greater detail potential additional interactions within the ALR2 inhibitor binding site. The experimental data obtained supported by molecular modeling indicated that inclusion of an N(2) substituent into the oxotriazinoindole scaffold of the parent drug OTI may increase ALR2 inhibition efficacy, e.g. about twice for the most efficient N(2)-CH2CH2COOH derivative 14, yet at the expense of inhibition selectivity relative to ALR1. The major drop in selectivity was explained by multiple interactions of the N(2) substituent with the residues of Arg309, Arg312 and Met302 located in the additional C-terminal loop of ALR1 missing in ALR2. Estimated passive gastrointestinal absorption of tested compounds is low and it could be improved by masking their carboxylic group (esterification or isosteric replacements).
Experimental section
Computational methods
For the in silico study, we used the method described in ref. [5]. The structures of human recombinant enzyme AKR1B1 in the complex with NADP+ and cemtirestat (CMTI) (PDB: 4QX4) [1] and the structure of porcine aldehyde reductase with 5-arylidene-2,4-thiazolidinedione (PDB: 3FX4) were used for docking. The used protocol has been already validated by a cross-docking procedure and confirmed a high value of accuracy in 90.0% [24].
Chemistry
Commercially available starting substances were purchased from Sigma-Aldrich (St. Louis, USA), Fluorochem (Hadfield, UK), AlfaAesar or Acros (parts of Thermo Fisher Scientific, UK or Belgium) vendors. Other chemicals and solvents were purchased from local sources with analytical grade quality. Merck Silica gel 60 F254 was used during the reaction by TLC analysis and for visualization of UV lamp (254 nm). 1H and 13C-NMR spectra were recorded on Varian Gemini instrument (600 and 150 MHz, resp.). Trimethylsilane (TMS) was used as an internal standard, chemical shifts are given in parts per million (ppm) and DMSO-d6 were used as a solvent. IR spectra were measured on an Agilent Technologies Cary 630 FTIR instrument with an MTS detector and a diamond probe. Melting points were measured on a Digital Melting Point Apparatus IA9200 and were noncorrected. Purity of compounds was performed on liquid chromatography - mass spectrometry (LC-MS; Agilent Technologies 1200 Series with Mass spectrometer Agilent Technologies 6100 Quadrupole LC-MS). All newly prepared and tested compounds 5, 8, 11 and 14 possess purity more than 95%.
General procedure
Appropriate ethyl ester 4, 7, 10 or 13 was dissolved in aqueous solution of NaOH. Reaction mixture was stirred at 80° C for 1 h. Complete conversion of starting material was confirmed by TLC analysis (SiO2, MeOH/EtOAc = 1/1). Afterwards the mixture was cooled in an ice bath and acidified with 1 M aq HCl solution to pH = 2. Obtained precipitate of product was filtered off and purified by crystallization from a mixture of H2O/DMSO. Yellow crystals were isolated and dried under reduced pressure to yield of product.
2-(3-Oxo-2-(2-oxopropyl)-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid (5)
Yellow crystals were isolated to yield 15.0 mg (0.050 mmol, 82%) of compound 5. Melting point: 259.1–264.8 °C [H2O / DMSO]. 1H-NMR (600 MHz, DMSO-d6): δ 13.17 (br s, 1H, -COOH), 7.92 (dd, 1H, J(8,9) = 7.6 Hz, J(7,9) = 1.4 Hz, H-C(9)), 7.62 (ddd, 1H, J(7,8) = 8.2 Hz, J(6,7) = 7.8 Hz, J(7,9) = 1.4 Hz, H-C(7)), 7.59 (dd, 1H, J(6,7) = 7.8 Hz, J(6,8) = 1.4 Hz, H-C(6)), 7.35 (ddd, 1H, J(7,8) = 8.2 Hz, J(8,9) = 7.6 Hz, J(6,8) = 1.4 Hz, H-C(8)), 5.12 and 4.95 (2x s, 2x2H, -CH2COOEt and -CH2COCH3), 2.25 (s, 3H, -CH2COCH3). 13C-NMR (150 MHz, DMSO-d6): δ 201.9 (-CH2COCH3), 168.6 (-CH2COOH), 153.7, 153.1, 143.9, 132.2, 131.1, 123.3, 121.1, 117.1, 111.4, 62.1, 42.1, 27.3 (-CH2COCH3). FT-IR (solid, cm−1): 3245–2080 (br, =CH-, -CH- and -COOH), 1740 (s), 1608 (s), 1575 (s), 1501 (s), 1465 (s), 1412 (s), 1370 (m), 1268 (m), 1218 (s), 1166 (s), 1088 (s), 1005 (s), 914 (s), 892 (m), 787 (s), 748 (s), 671 (m). MS (ESI m/z, negative ion mode): 299.0 (32%) [M-H]+, 255.1 (67%) [M-CO2-H]+. Elemental analysis. Anal. Calcd for C14H12N4O4 (300.27): C, 56.00; H, 4.03; N, 18.66; found: C, 56.31; H, 4.30; N, 18.45.
Ethyl 2-(2-(2-acetoxyethyl)-3-oxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetate (8)
Yellow crystals were isolated to yield 55.7 mg (0.19 mmol, 69%) of compound 8. Melting point: 256.4–260.7 °C [H2O / DMSO]. 1H-NMR (600 MHz, DMSO-d6): δ 13.32 (br s, 1H, -CH2COOH), 7.95 (dd, 1H, J(8,9) = 7.6 Hz, J(7,9) = 1.0 Hz, H-C(9)), 7.60 (ddd, 1H, J(7,8) = 7.9 Hz, J(6,7) = 7.7 Hz, J(7,9) = 1.0 Hz, H-C(7)), 7.57 (dd, 1H, J(6,7) = 7.7 Hz, J(6,8) = 1.0 Hz, H-C(6)), 7.33 (ddd, 1H, J(7,8) = 7.9 Hz, J(8,9) = 7.6 Hz, J(6,8) = 1.0 Hz, H-C(8)), 4.93 (s, 2H, -CH2-N(5)), 4.86 (br s, 1H, -CH2CH2OH), 4.24 (t, 2H, J(CH2,CH3) = 5.5 Hz, -N(2)CH2CH2OH), 3.80 (t, 2H, J(CH2,CH3) = 5.5 Hz, -N(2)CH2CH2OH). 13C-NMR (150 MHz, DMSO-d6): δ 168.7 (-NCH2COOH), 154.3, 153.3, 143.7, 131.5, 130.7, 123.1, 120.9, 117.3, 111.2, 58.3, 55.8, 42.1. FT-IR (solid, cm−1): 2605 (w), 2344 (w), 1731 (m), 1680 (m), 1601 (s), 1589 (s), 1504 (m), 1470 (m), 1401 (s), 1338 (s), 1276 (m), 1144 (m), 1118 (m), 1688 (w), 941 (m), 790 (m), 750 (s), 549 (m), 511 (m), 438 (s). MS (ESI m/z, negative ion mode): 287.0 (82%) [M-H]-, 243.0 (13%) [M-CO2-H]-. Elemental analysis. Anal. Calc for C13H12N4O4 (288.26): C, 54.17; H, 4.20; N, 19.44; found: C, 54.53; H, 4.12; N, 19.35.
2-(2-(2-Methoxyethyl)-3-oxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid (11)
Yellow crystals were isolated to yield 50.1 mg (0.17 mmol, 94%) of acid 11. Melting point: 230.8–235.6 °C [H2O/HCl]. 1H-NMR: (600 MHz, DMSO-d6): δ 13.30 (br s, 1H, -COOH), 7.95 (dd, 1H, J(8,9) = 7.5 Hz, J(7,9) = 1.0 Hz, H-C(9)), 7.61 (ddd, 1H, J(7,8) = 7.8 Hz, J(6,7) = 7.6 Hz, J(7,9) = 1.0 Hz, H-C(7)), 7.58 (dd, 1H, J(6,7) = 7.6 Hz, J(6,8) = 1.0 Hz, H-C(6)), 7.34 (ddd, 1H, J(7,8) = 7.8 Hz, J(8,9) = 7.5 Hz, J(6,8) = 1.0 Hz, H-C(8)), 4.93 (s, 2H, -CH2-COOH), 4.35 and 3.75 (2 x t, 2 x 2H, 2 x J(CH2,CH3) = 5.4 Hz, -N(2)CH2CH2OCH3) and N(2)CH2CH2OCH3), 3.25 (s, 3H, -OCH3). 13C-NMR: (150 MHz, DMSO-d6): δ 168.7 (-COOH), 153.4, 153.1, 143.8, 131.7, 130.9, 123.1, 121.0, 117.2, 111.2, 68.8, 57.9, 52.4, 42.0. FT-IR (solid, cm−1): 3244 (w), 2918 (w), 1741 (s), 1607 (s), 1576 (s), 1464 (m), 1411 (m), 1308 (m), 1266 (m), 1218 (s), 1166 (s), 1088 (s), 1006 (s), 914 (s), 891 (m), 757 (s), 747 (s), 670 (m). MS (ESI m/z, negative ion mode): 301.1 (100%) [M-H]−, 257.1 (47%) [M-CO2-H]−. Elemental analysis. Anal. Calc for C14H14N4O4 (302.29): C, 55.63; H, 4.67; N, 18.53 found: C, 55.53; H, 4.49; N, 18.79.
3-(5-(Carboxymethyl)-3-oxo-3,5-dihydro-2H-[1,2,4]triazino[5,6-b]indol-2-yl)propanoic acid (14)
Precipitated product 14 was filtered off and purified by crystallization from a mixture of H2O / HCl. Yellow crystals were isolated by filtration and dried under high vacuum to yield 50.1 mg (0.16 mmol, 73%) of compound 14. Compound 14 was not yet described in the literature. Melting point: 273.0–277.4 °C [H2O/HCl]. 1H-NMR: (600 MHz, DMSO-d6): δ 12.53 (2 x br s, 2 x 1H, 2 x -COOH), 7.93 (d, 1H, J(8,9) = 6.5 Hz, J(7,9) = 0.9 Hz, H-C(9)); 7.60 (dd, 1H, J(7,8) = 7.0 Hz, J(6,7) = 6.9 Hz, J(7,9) = 0.9 Hz, H-C(7)); 7.57 (d, 1H, J(6,7) = 6.9 Hz, J(6,8) = 0.9 Hz, H-C(6)); 7.34 (dd, 1H, J(7,8) = 7.0 Hz, J(8,9) = 6.5 Hz, J(6,8) = 0.9 Hz, H-C(8)); 4.93 (s, 2H, -N(5)CH2COOH), 4.37 (t, 2H, J(CH2,CH2) = 7.0 Hz, -N(2)CH2CH2COOH); 2.80 (t, 2H, J(CH2,CH2) = 7.0 Hz, -N(2)CH2CH2COOH). 13C-NMR: (150 MHz, DMSO-d6): δ 172.2, 168.7, 153.4, 152.9, 143.9, 131.6, 130.9, 123.1, 120.9, 117.2, 111.3, 49.1, 42.0, 32.4. FT-IR (solid, cm−1): 2924 (m), 2716 (w), 2598 (w), 2524 (w),1724 (s), 1641 (m), 1596 (m), 1596 (m), 1562 (m), 1502 (m), 1489 (m), 1406 (m), 1373 (s), 1324 (m), 1206 (s), 1134 (m), 1012 (m), 909 (m), 787 (s), 751 (s), 689 (w), 661 (w), 599 (w), 490 (w), 434 (w) cm−1. MS (ESI m/z, positive ion mode): 317.1 [M + H]+ (100%). Elemental analysis. Anal. Calc for C14H12N4O5 (316.27): C, 53.17; H, 3.82; N, 17.72; found: C, 53.05; H, 3.69; N, 17.59.
All final products 5, 8, 11, and 14 also with intermediates (see Scheme 1) are described in more details of the Supporting Information to this paper.
Interference compounds assay
Compounds 5, 8, 11, and 14 underwent screening using three in silico tools to eliminate potential false positives with nonspecific interactions (PAINS) and aggregation-forming capabilities. The screening tools utilized were http://advisor.docking.org, http://www.swissadme.ch/ and http://zinc15.docking.org/patterns/home. All the compounds successfully passed these filters, and no structural alerts were identified.
Animals
Male Wistar rats, aged 8–9 weeks and weighing between 200 and 230 g, served as organ donors. These animals originated from the Breeding Facility of the Institute of Experimental Pharmacology in Dobrá Voda, Slovak Republic. The study received approval from the Ethics Committee of the Institute and adhered to the Principles of Laboratory Animal Care (NIH publication 83-25, revised 1985) and Slovak laws governing animal experiments (Decree 289, Part 139, July 9th, 2003).
Enzyme assays
The procedure for preparing enzymes of ALR2 from rat lenses and ALR1 from rat kidneys has been documented in previously reports. Spectrophotometric assays were employed to measure enzyme activities, specifically by quantifying NADPH consumption at 340 nm [25]. Comprehensive details of the experimental methods can be found in the Supporting Information accompanying this paper.
Estimation of passive gastrointestinal absorption
Passive gastrointestinal absorption of tested compounds (5, 8, 11 and 14) and standards (OTI, OTI−1, CMTI and epalrestat) was estimated using biopartitioning micellar chromatography. The analysis was performed on Agilent 1200 HPLC chromatograph (Agilent Technologies, Palo Alto, CA, USA), which consisted of binary pump, manual injector (injection volume: 20 μL) and PDA detector. Zorbax Extend-C18 column (150 mm × 4.6 mm, 5 μm particle size; Agilent Technologies, Palo Alto, CA, USA) was used. The mobile phase was prepared by mixing aqueous phase (40 mM solution of Brij35 (Sigma Aldrich, Steinheim, Germany) prepared in 7 mM disodium hydrogen phosphate (Merck, Darmstadt, Germany)) with acetonitrile (Fisher, Loughborough, UK). The aqueous phase/acetonitrile ratio was 95:5 (v/v). The mobile phase pH was adjusted to 3.0 or 5.5 by addition of phosphoric acid (Merck, Darmstadt, Germany). The temperature of the column was 36.5 °C, the flow rate was set to 1 mL/min and chromatographic peaks were detected at 230 nm (dimethyl sulfoxide) and 260 nm (tested compounds). Stock solutions of tested compounds were prepared in dimethyl sulfoxide (Fisher, Loughborough, UK) to obtain concentration 2 mg/ml. To prepare working solutions (0.02 mg/ml), stock solutions were diluted with the mobile phase. Finally, retention times of analytes (tR) and dimethyl sulfoxide (t0) were determined and corresponding retention factors (k) were calculated (Eq.(1)).
Data availability
Data will be made available from the corresponding author on reasonable request.
References
Stefek M, Prnova MS, Majekova M, Rechlin C, Heine A, Klebe G. Identification of Novel Aldose Reductase Inhibitors Based on Carboxymethylated Mercaptotriazinoindole Scaffold. J Med Chem. 2015;5:2649–57. https://doi.org/10.1021/jm5015814.
Stefek M, Milackova I, Diez-Dacal B, Pérez-Sala D, Soltesova Prnova M. Use of 5-carboxymethyl-3-mercapto−1,2,4-triazino-[5,6-b]indoles and their pharmaceutical composition. WO2015/057175, 3.4.2015.
Prnova MS, Ballekova J, Majekova M, Stefek M. Antioxidant Action of 3-Mercapto-5H−1,2,4-Triazino[5,6-b]Indole-5-Acetic Acid, an Efficient Aldose Reductase Inhibitor, in a 1,1′-Diphenyl-2-Picrylhydrazyl Assay and in the Cellular System of Isolated Erythrocytes Exposed to Tert-Butyl Hydroperoxide. Redox Rep. 2015;20:282–8. https://doi.org/10.1179/1351000215Y.0000000019.
Maccari R, Ottana R. Targeting aldose reductase for the treatment of diabetes complications and inflammatory diseases: new insights and future directions. J Med Chem. 2015;58:2047–67. https://doi.org/10.1021/jm500907a.
Prnova MS, Medina-Campos O, Pedraza-Chaverri J, Colín-González AL, Piedra-García F, Rangel-López E, et al. Antioxidant Mechanisms in the Neuroprotective Action of Cemtirestat: Studies in Chemical Models, Liposomes and Rat Brain Cortical Slices. Neurosci. 2020;443:206–17. https://doi.org/10.1016/j.neuroscience.2020.07.014.
Hlaváč M, Kováčiková L, Prnová MŠ, Šramel P, Addová G, Májeková M, et al. Development of Novel Oxotriazinoindole Inhibitors of Aldose Reductase: Isosteric Sulfur/Oxygen Replacement in the Thioxotriazinoindole Cemtirestat Markedly Improved Inhibition Selectivity. J Med Chem. 2020;63:369–81. https://doi.org/10.1021/acs.jmedchem.9b01747.
Hlaváč M, Kováčiková L, Prnová MŠ, Addová G, Hanquet G, Štefek M, et al. Novel Substituted N-Benzyl(Oxotriazinoindole) Inhibitors of Aldose Reductase Exploiting ALR2 Unoccupied Interactive Pocket. Bioorg Med Chem. 2020;29:115885 https://doi.org/10.1016/j.bmc.2020.115885.
Barski OA, Gabbay KH, Bohren KM. The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity. Biochemistry. 1996;35:14276–80. https://doi.org/10.1021/bi9619740.
Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40:553–624. https://doi.org/10.1080/03602530802431439.
Rees-Milton KJ, Jia Z, Green NC, Bhatia M, El-Kabbani O, Flynn TG. Aldehyde reductase: the role of C-terminal residues in defining substrate and cofactor specificities. Arch Biochem Biophys. 1998;15:137–44. https://doi.org/10.1006/abbi.1998.0721.
Steuber H, Heine A, Podjarny A, Klebe G. Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features. J Mol Biol. 2008;379:991–1016. https://doi.org/10.1016/j.jmb.2008.03.063.
El-Kabbani O, Old SE, Ginell SL, Carper DA. Aldose and aldehyde reductases: structure-function studies on the coenzyme and inhibitor-binding sites. Mol Vis. 1999;5:20.
Majekova M. Ligand-based drug design of novel aldose reductase inhibitors. Fut Med Chem. 2018;10:2493–6. https://doi.org/10.4155/fmc-2018-0127.
Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–1. https://doi.org/10.1016/S1359-6446(04)03069-7.
Abad-Zapatero C. Ligand efficiency indices for effective drug discovery. Expert Opin Drug Discov. 2007;2:469–88. https://doi.org/10.1517/17460441.2.4.469.
Keserü GM, Makara GM. The Influence of Lead Discovery Strategies on the Properties of Drug Candidates. Nat Rev Drug Discov. 2009;8:203–12. https://doi.org/10.1038/nrd2796.
Hopkins AL, Keserü GM, Leeson PD, Rees DC, Reynolds CH. The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov. 2014;13:105–121. https://doi.org/10.1038/nrd4163.
Perola E. An Analysis of the Binding Efficiencies of Drugs and their Leads in Successful Drug Discovery Programs. J Med Chem. 2010;53:2986–97.
Escuder-Gilabert L, Martınez-Pla JJ, Sagrado S, Villanueva-Camañas RM, Medina-Hernández MJ. Biopartitioning micellar separation methods: modelling drug absorption. J Chromat B. 2023;797:21–35. https://doi.org/10.1016/s15700232(03)00606-8.
Dobričić V, Savić J, Nikolic K, Vladimirov S, Vujić Z, Brborić J. Application of biopartitioning micellar chromatography and QSRR modeling for prediction of gastrointestinal absorption and design of novel β-hydroxy-β-arylalkanoic acids. Eur J Pharm Sci. 2017;100:280–4. https://doi.org/10.1016/j.ejps.2017.01.023.
Marieb EN, Hoehn K. Human Anatomy and Physiology. 8th ed. San Francisco, CA, USA: Benjamin Cummings; 2010, 32, p.28.
Ovesen L, Bendtsen F, Tage-Jensen U, Pedersen NT, Gram BR, Rune SJ. Intraluminal pH in the stomach, duodenum, and proximal jejunum in normal subjects and patients with exocrine pancreatic insufficiency. Gastroenterology. 1986;90:958–62. https://doi.org/10.1016/0016-5085(86)90873-5.
Maurer JM, Schellekens RC, Van Rieke HM, Wanke C, Iordanov V, Stellaard F, et al. Gastrointestinal pH and transit time profiling in healthy volunteers using the IntelliCap system confirms ileo-colonic release of ColoPulse tablets. PLoS One. 2015;10:e0129076 https://doi.org/10.1371/journal.pone.0129076.
Majekova M, Ballekova J, Prnova M, Stefek M. Structure Optimization of Tetrahydropyridoindole-Based Aldose Reductase Inhibitors Improved their Efficacy and Selectivity. Bioorg Med Chem. 2017;25:6353–6360. https://doi.org/10.1016/j.bmc.2017.10.005.
Stefek M, Snirc V, Djoubissie PO, Majekova M, Demopoulos V, Rackova L, et al. Carboxymethylated pyridoindole antioxidants as aldose reductase inhibitors: synthesis, activity, partitioning, and molecular modeling. Bioorg Med Chem. 2008;16:4908–20. https://doi.org/10.1016/j.bmc.2008.03.039.
Kumar M, Choudhary S, Singh PK, Silakari O. Addressing selectivity issues of aldose reductase 2 inhibitors for the management of diabetic complications. Future Med Chem. 2020;12:1327–58. https://doi.org/10.4155/fmc-2020-0032.
Acknowledgements
This work was supported by VEGA 2/0008/22, VEGA 2/0103/22, APVV-20-0411, APVV-20-0543, APVV SK-SRB-21-0047 (451-03-43/2022-09/02) by the Ministry of Science, Technological Development and Innovation, Republic of Serbia through Grant Agreement with University of Belgrade-Faculty of Pharmacy, APVV SK-FR-22-0017 and by the Operation Program of Integrated Infrastructure for the project, Advancing University Capacity and Competence in Research, Development and Innovation (Accord), ITMS2014+: 313021×329 and ITMS2014+: 313021BUZ3 (Usccord) co-financed by the European Regional Development Fund.
Author contributions
MS, AB and LK designed experiments, wrote and critically reviewed the manuscript; AB designed novel OTI inhibitors; AA, KA and GH performed or supported the chemical syntheses; SG responsible for checking the correctness of the syntheses; GA performed spectral and purity analysis; LK and KA performed enzyme preparations and enzyme assays; MM carried out computer molecular modeling and selectivity studies; LK performed calculations of physicochemical properties and parameters of molecular obesity and PAINS screening. VD performed estimation of passive gastrointestinal absorption using biopartitioning micellar chromatography.
Funding
Open access funding provided by The Ministry of Education, Science, Research and Sport of the Slovak Republic in cooperation with Centre for Scientific and Technical Information of the Slovak Republic.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary infomation
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kováčiková, L., Gaikwad, S., Almášiová, K. et al. Selectivity of N(2)-substituted oxotriazinoindole aldose reductase inhibitors is determined by the interaction pattern with Pro301-Arg312 loop of aldehyde reductase. Med Chem Res 33, 492–503 (2024). https://doi.org/10.1007/s00044-024-03194-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-024-03194-3