Abstract
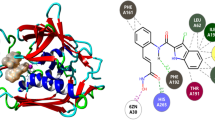
Leptospirosis is a worldwide waterborne zoonosis caused by Leptospira interrogans serovar lai strain 56601 and no specific antibiotics are available to effectively treat the disease to date. The enzyme 3-methyladenine-DNA glycosylase I (LiTagA), which excises 3-methyadenine and 3-methylguanine from alkylated DNA, is an indispensable component for the survival of the bacteria strain, and the absence of the enzyme in human makes it a promising chemotherapeutic target for antileptospiral treatments. In the present study, 578 non-redundant analogs of a few numbers of 3-alkyladenines, experimentally reported inhibitors to the enzyme, were retrieved from the NCI database and their binding affinities on the enzyme (modeled, solvated, and energy minimized at close quarters of physiologic conditions) were screened by means of PyRx—a high throughput virtual screening tool—and AutoDock. Using a lead identified from the studies as a seed, a small focussed combinatorial library containing 200 de novo inhibitors to the enzyme was generated and the compounds were subjected to affinity filter, bioavailability filter, and toxicity filter by means of an array of computational tools. Combined comprehensive analysis on the data obtained from the various filters brought out few numbers of efficient de novo inhibitors to the Leptospira interrogans LiTagA enzyme and also revealed that 5-H-indeno[1,2-b]pyridine system flanked by cationic and anionic chemical moieties at sites 9th and 12th positions, respectively, is an essential skeleton for the highly potent inhibitors to the enzyme. Moreover, implications of the studies on designing drug-likeness de novo inhibitors to protein targets have also been discussed in detail.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR (2001) The InterPro database: an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29:37–40
Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer ELL (2000) The Pfam protein families database. Nucleic Acids Res 28:263–266
Bjelland S, Bjoras M, Seeberg E (1993) Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res 21:2045–2049
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Deleage G, Blanchet C, Geourjon C (1997) Protein structure prediction. Implications for the biologist. Biochimie 79(11):681–686
Drohat AC, Kwon K, Krosky DJ, Stivers JT (2002) 3-Methyladenine DNA glycosylase I is an unexpected helix-hairpin-helix superfamily member. Nat Struct Biol 9:659–664
Elżbieta S, Jarosław MC, Maria AG, Laval J, Zygmunt K, Barbara T (2005) Inhibition of DNA repair glycosylases by base analogs and tryptophan pyrolysate, Trp-P-1. Acta Biochim Pol 52:167–178
Faine S, Adler B, Bolin C, Perolat P (2000) Leptospira and leptospirosis, 2nd edn. Medisci Press, Melbourne
Francisco M, Sanchez R, Sali A (2002) Statistical potentials for fold assessment. Protein Sci 11:430–448
Fronza G, Gold BJ (2004) The biological effects of N3-methyladenine. Cell Biochem 91:250–257
Gill SC, Von Hippel PH (1989) Extinction coefficient. Anal Biochem 182:319–328
Guruprasad K, Reddy BVP, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 4:155–164
Hess B (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Ikai AJ (1980) Thermo stability and aliphatic index of globular proteins. J Biochem 88:1895–1898
Kyte J, Doolottle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Laurie AT, Jackson RM (2005) Q-SiteFinder: an energy-based method for the prediction of protein–ligand binding sites. Bioinformatics 21:1908–1916
Laval J (1996) Role of DNA repair enzymes in the cellular resistance to oxidative stress. Pathol Biol 44:14–24
Levett PN (2001) Leptospirosis. Clin Microbiol 14:296–326
Maria AM, Stephanie V, Matthieu M, David G, Pierre T, Bruno OV (2006) FAF-Drugs: free ADME/tox filtering of compound collections. Nucleic Acids Res 34:738–744
Metz AH, Hollis T, Eichman BF (2007) DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG). EMBO J 26(9):2411–2420
Moe E, Hall DR, Leiros I, Monsen VT, Timmins J, McSweeney S (2012) Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans. Acta Crystallogr D Biol Crystallogr 68:703–712
Morris GM (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Murvai J, Vlahovicek K, Barta E, Pongor S (2001) The SBASE protein domain library, release 8.0: a collection of annotated protein sequence segments. Nucleic Acids Res 29:58–60
O’Connor TR (1993) Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res 21:5561–5569
Ren SX, Fu G (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28:231–234
Schüttelkopf AW, van Aalten DMF (2004) PRODRG-a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr 60:1355–1363
Tudek B, VanZeeland AA, Kusmierek JT, Laval J (1998) Activity of Escherichia coli DNA-glycosylases on DNA damaged by methylating and ethylating agents and influence of 3-substituted adenine derivatives. Mutat Res 407:169–176
Vijayachari P, Sugunan AP, Shriram AN (2008) Leptospirosis: an emerging global public health problem. J Biosci 33:557–569
Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12:1073–1086
Wang R, Gao Y, Lai L (2000) LigBuilder: a multi-purpose program for structure-based drug design. J Mol Model 6:498–516
Wang Z, Jin L, Wegrzyn A (2007) Leptospirosis vaccines. Microb Cell Fact 6:39–48
Wolf LK (2009) New software and websites for the chemical enterprise. Chem Eng News 8:7–31
Xiaofeng Z, Xuan Y, Lester GC, Huanting L, Shirley G, Peter JC, James N (2012) A model for 3-methyladenine recognition by3-methyladenine DNA glycosylase I (TAG) from Staphylococcus aureus. Acta Crystallogr 68:610–615
Acknowledgments
The authors thank Ms. N. Lavanya Roseline for her contributions on the earlier stage of this research project. The authors would also like to acknowledge the “Centre of Excellence”—AYUSH at SASTRA University, for generously allowing us to use Schrödinger software suite supported by the AYUSH research Grant (Z.15015/01/2010), Government of India.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rajesh, S.S., Sivaraman, T. Cheminformatic designing of de novo inhibitors to 3-methyl adenine DNA glycosylase I (LiTagA) from Leptospira interrogans serovar lai strain 56601 . Med Chem Res 22, 3434–3443 (2013). https://doi.org/10.1007/s00044-012-0346-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-012-0346-x