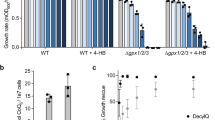
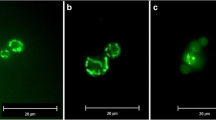
Abstract
The Voltage-Dependent Anion-selective Channel (VDAC) is the pore-forming protein of mitochondrial outer membrane, allowing metabolites and ions exchanges. In Saccharomyces cerevisiae, inactivation of POR1, encoding VDAC1, produces defective growth in the presence of non-fermentable carbon source. Here, we characterized the whole-genome expression pattern of a VDAC1-null strain (Δpor1) by microarray analysis, discovering that the expression of mitochondrial genes was completely abolished, as consequence of the dramatic reduction of mtDNA. To overcome organelle dysfunction, Δpor1 cells do not activate the rescue signaling retrograde response, as ρ0 cells, and rather carry out complete metabolic rewiring. The TCA cycle works in a “branched” fashion, shunting intermediates towards mitochondrial pyruvate generation via malic enzyme, and the glycolysis-derived pyruvate is pushed towards cytosolic utilization by PDH bypass rather than the canonical mitochondrial uptake. Overall, Δpor1 cells enhance phospholipid biosynthesis, accumulate lipid droplets, increase vacuoles and cell size, overproduce and excrete inositol. Such unexpected re-arrangement of whole metabolism suggests a regulatory role of VDAC1 in cell bioenergetics.







Similar content being viewed by others
References
Messina A, Reina S, Guarino F, De Pinto V (2012) VDAC isoforms in mammals. Biochim Biophys Acta Biomembr 1818:1466–1476
Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N (2010) VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Asp Med 31:227–285
Bayrhuber M, Menis T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K (2008) Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA 105:15370–15375
Hiller S, Graces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210
Ujwal R, Cascio D, Colletier JP, Fahama S, Zhanga J, Torod L, Pinga P, Abramson J (2008) The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA 105:17742–17747
Schredelseker J, Paz A, López CJ, Altenbach C, Leung CS, Drexler MK, Chen JN, Hubbell WL, Abramson J (2014) High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J Biol Chem 289:12566–12577
Magrì A, Messina A (2017) Interactions of VDAC with proteins involved in neurodegenerative aggregation: an opportunity for advancement on therapeutic molecules. Curr Med Chem 24:4470–4487
Magrì A, Reina S, De Pinto V (2018) VDAC1 as pharmacological target in cancer and neurodegeneration: focus on its role in apoptosis. Front Chem 6:108
Reina S, Guarino F, Magrì A, De Pinto V (2016) VDAC3 as a potential marker of mitochondrial status is involved in cancer and pathology. Front Oncol 6:264
Reina S, De Pinto V (2017) Anti-cancer compound targeted to VDAC: potential and perspectives. Curr Med Chem 24:4447–4469
Ellenrieder L, Dieterle MP, Nguyen Doan K, Mårtensson CU, Floerchinger A, Campo ML, Pfanner N, Becker T (2019) Dual role of mitochondrial porin in metabolite transport across the outer membrane and protein transfer to the inner membrane. Mol Cell 73:1056–1065
Dihanich M, Suda K, Schatz G (1987) A yeast mutant lacking mitochondrial porin is respiratory-deficient, but can recover respiration with simultaneous accumulation of an 86-kd extramitochondrial protein. EMBO J 6:723–728
Dihanich M, Schmid A, Oppliger W, Benz R (1989) Identification of a new pore in the mitochondrial outer membrane of a porin-deficient yeast mutant. Eur J Biochem 182:703–708
Blachly-Dyson E, Song J, Wolfgang WJ, Colombini M, Forte M (1997) Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Mol Cell Biol 17:5727–5738
Guardiani C, Magrì A, Karachitos A, Di Rosa MC, Reina S, Bodrenko I, Messina A, Kmita H, Ceccarelli M, De Pinto V (2018) yVDAC2, the second mitochondrial porin isoform of Saccharomyces cerevisiae. Biochim Biophys Acta Bioenerg 1859:270–279
Magrì A, Karachitos A, Di Rosa MC, Reina S, Conti Nibali S, Messina A, Kmita H, De Pinto V (2019) Recombinant yeast VDAC2: a comparison of electrophysiological features with the native form. FEBS Open Bio 9:1184–1193
Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Höß P, Feuerstein R, Gebert M, Bohnert M, van der Laan M, Schuldiner M, Schütze C, Oeljeklaus S, Pfanner N, Wiedemann N, Warscheid B (2017) Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19:2836–2852
Orlandi I, Pellegrino Coppola D, Vai M (2014) Rewiring yeast acetate metabolism through MPC1 loss of function leads to mitochondrial damage and decreases chronological lifespan. Microbial Cell 1:393–405
Leggio L, Guarino F, Magrì A, Accardi-Gheit R, Reina S, Specchia V, Damiano F, Tomasello MF, Tommasino M, Messina A (2018) Mechanism of translation control of the alternative Drosophila melanogaster voltage dependent anion channel 1 mRNAs. Sci Rep 8:5347
Löke M, Kristjuhan K, Kristjuhan A (2011) Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50:325–328
Livak KJ, Schmittgen TD (2011) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Koning AJ, Lum PY, Williams JM, Wright R (1993) DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskelet 25:111–128
Magrì A, Belfiore R, Reina S, Tomasello MF, Di Rosa MC, Guarino F, Leggio L, De Pinto V, Messina A (2016) Hexokinase I N-terminal based peptide prevents the VDAC1-SOD1 G93A interaction and re-establishes ALS cell viability. Sci Rep 6:34802
Orlandi I, Ronzulli R, Casatta N, Vai M (2013) Ethanol and acetate acting as carbon/energy sources negatively affect yeast chronological aging. Oxid Med Cell Longev 2013:802870
Orlandi I, Pellegrino Coppola D, Strippoli M, Ronzulli R, Vai M (2017) Nicotinamide supplementation phenocopies SIR2 inactivation by modulating carbon metabolism and respiration during yeast chronological aging. Mech Ageing Dev 161:277–287
Orlandi I, Stamerra G, Vai M (2018) Altered expression of mitochondrial NAD+ carriers influences yeast chronological lifespan by modulating cytosolic and mitochondrial metabolism. Front Genet 9:676
Trindade D, Pereira C, Chaves SR, Manon S, Côrte-Real M, Sousa MJ (2016) VDAC regulates AAC-mediated apoptosis and cytochrome c release in yeast. Microb Cell 3:500–510
Aranda A, del Olmo M (2003) Response to acetaldehyde stress in the yeast Saccharomyces cerevisiae involves a strain-dependent regulation of several ALD genes and is mediated by the general stress response pathway. Yeast 20:747–759
Postma E, Verduyn C, Scheffers WA, Van Dijken JP (1989) Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 55:468–477
de Assis LJ, Zingali RB, Masuda CA, Rodrigues SP, Montero-Lomelí M (2013) Pyruvate decarboxylase activity is regulated by the Ser/Thr protein phosphatase Sit4p in the yeast Saccharomyces cerevisiae. FEMS Yeast Res 13:518–528
Steensma HY, Tomaska L, Reuven P, Nosek J, Brandt R (2008) Disruption of genes encoding pyruvate dehydrogenase kinases leads to retarded growth on acetate and ethanol in Saccharomyces cerevisiae. Yeast 25:9–19
Bourque SD, Titorenko VI (2009) A quantitative assessment of the yeast lipidome using electrospray ionization mass spectrometry. J Vis Exp 30:1513
Gallet PF, Maftah A, Petit JM, Denis-Gay M, Julien R (1995) Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur J Biochem 228:113–119
Greenberg ML, Reiner B, Henry SA (1982) Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics 100:19–33
Vanoni M, Vai M, Popolo L, Alberghina L (1983) Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J Bacteriol 156:1282–1291
Stuart RA (2008) Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr 40:411–417
Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W (2012) Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem 287:23095–23103
Moran JV, Mecklenburg KL, Sass P, Belcher SM, Mahnke D, Lewin A, Perlman P (1994) Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: initial definition of the maturase domain of the group II intron aI2. Nucleic Acids Res 22:2057–2064
Magrì A, Di Rosa MC, Tomasello MF, Guarino F, Reina S, Messina A, De Pinto V (2016) Overexpression of human SOD1 in VDAC1-less yeast restores mitochondrial functionality modulating beta-barrel outer membrane protein genes. Biochim Biophys Acta Bioenerg 1857:789–798
Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD (2005) Sir2 blocks extreme life-span extension. Cell 123:655–667
Pronk JT, Yde Steensma H, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Bender T, Pena G, Martinou JC (2015) Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J 34:911–924
Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, Rutter J (2012) A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337:96–100
Butow RA, Avadhani NG (2004) Mitochondrial signaling: the retrograde response. Mol Cell 14:1–15
Epstein CB, Waddle JA, Hale W 4th, Davé V, Thornton J, Macatee TL, Garner HR, Butow RA (2001) Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell 12:297–308
de Smidt O, du Preez JC, Albertyn J (2008) The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res 8:967–978
Saint-Prix F, Bönquist L, Dequin S (2004) Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology 150:2209–2220
van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY (1996) The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem 271:28953–28959
Franken J, Kroppenstedt S, Swiegers JH, Bauer FF (2008) Carnitine and carnitine acetyltransferases in the yeast Saccharomyces cerevisiae: a role for carnitine in stress protection. Curr Genet 53:347–360
van Roermund CW, Hettema EH, van den Berg M, Tabak HF, Wanders RJ (1999) Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J 18:5843–5852
Tehlivets O, Scheuringer K, Kohlwein SD (2007) Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta Mol Cell Biol Lipids 1771:255–270
Vahlensieck HF, Pridzun L, Reichenbach H, Hinnen A (1994) Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet 25:95–100
Hofbauer HF, Schopf FH, Schleifer H, Knittelfelder OL, Pieber B, Rechberger GN, Wolinski H, Gaspar ML, Kappe CO, Stadlmann J, Mechtler K, Zenz A, Lohner K, Tehlivets O, Henry SA, Kohlwein SD (2014) Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev Cell 29:729–739
Koch B, Schmidt C, Daum G (2014) Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol Rev 38:892–915
Graef M (2018) Lipid droplet-mediated lipid and protein homeostasis in budding yeast. FEBS Lett 592:1291–1303
Ren M, Phoon CK, Schlame M (2014) Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res 55:1–16
Carman GM, Han GS (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Ann Rev Biochem 80:859–883
Henry SA, Kohlwein SD, Carman GM (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190:317–349
Henry SA, Gaspar ML, Jesch SA (2014) The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast. Chem Phys Lipids 180:23–43
Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304:1644–1647
Han GS, Carman GM (2017) Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J Biol Chem 292:13230–13242
Becker L, Bannwarth M, Meisinger C, Hill K, Model K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, Wagner R (2005) Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol 353:1011–1020
Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N (2008) Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132:1011–1024
Kmita H, Antos N, Wojtkowska M, Hryniewiecka L (2004) Processes underlying the upregulation of tom proteins in S. cerevisiae mitochondria depleted of the VDAC channel. J Bioenerg Biomembr 36:187–193
Flinner N, Ellenrieder L, Stiller SB, Becker T, Schleiff E, Mirus O (2013) Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta Mol Cell Res 1833:3314–3325
Krüger V, Becker T, Becker L, Montilla-Martinez M, Ellenrieder L, Vögtle FN, Meyer HE, Ryan MT, Wiedemann N, Warscheid B, Pfanner N, Wagner R, Meisinger C (2017) Identification of new channels by systematic analysis of the mitochondrial outer membrane. J Cell Biol 216:3485–3495
Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quiñones JF, Jazwinski SM (2012) Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Front Genet 2:102
Yi DG, Hong S, Huh WK (2018) Mitochondrial dysfunction reduces yeast replicative lifespan by elevating RAS-dependent ROS production by the ER-localized NADPH oxidase Yno1. PLoS One 13:e0198619
Torelli NQ, Ferreira-Júnior JR, Kowaltowski AJ, da Cunha FM (2015) RTG1- and RTG2-dependent retrograde signalling controls mitochondrial activity and stress resistance in Saccharomyces cerevisiae. Free Rad Biol Med 81:30–37
Miner GE, Starr ML, Hurst LR, Fratti RA (2017) Deleting the DAG kinase Dgk1 augments yeast vacuole fusion through increased Ypt7 activity and altered membrane fluidity. Traffic 18:315–329
Starr ML, Fratti RA (2019) The participation of regulatory lipids in vacuole homotypic fusion. Trends Biochem Sci 44:546–554
Warburg O (1925) The metabolism of carcinoma cells. J Cancer Res 9:148–163
Shim H, Chun YS, Lewis BC, Dang CV (1998) A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA 95:1511–1516
Fantin VR, St-Pierre J, Leder P (2006) Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9:425–434
Liberti MV, Locasale JW (2006) The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41:211–218
Natter K, Kholwein SD (2016) Yeast and cancer cells—common principles in lipid metabolism. Biochim Biophys Acta Mol Cell Biol Lipids 1831:314–326
Macheda ML, Rogers S, Best JD (2005) Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol 203:654–662
Diaz-Ruiz R, Rigoulet M, Devin A (2011) The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta Bioenerg 1807:568–576
Bola O, Hampsey JJ, Hampsey M (2015) Genetic analysis of the Warburg effect in yeast. Adv Biol Regul 57:185–192
Raghavan A, Sheiko T, Graham BH, Craigen WJ (2012) Voltage-dependant anion channels: novel insights into isoform function through genetic models. Biochim Biophys Acta Biomembr 1818:1477–1485
Sampson MJ, Decker WK, Beaudet AL, Ruitenbeek W, Armstrong D, Hicks MJ, Craigen WJ (2011) Immotile sperm and infertility in mice lacking mitochondrial voltage-dependent anion channel type 3. J Biol Chem 276:39206–39212
Anflous-Pharayra K, Lee N, Armstrong DL, Craigen WJ (2011) VDAC3 has differing mitochondrial functions in two types of striated muscles. Biochim Biophys Acta Bioenerg 1807:150–156
Maldonado EN, Sheldon KL, DeHart DN, Patnaik J, Manevich Y, Townsend DM, Bezrukov SM, Rostovtseva TK, Lemasters JJ (2013) Voltage-dependent anion channels modulate mitochondrial metabolism in cancer cells: regulation by free tubulin and erastin. J Biol Chem 288:1192–11929
Naghdi S, Várnai P, Hajnóczky G (2015) Motifs of VDAC2 required for mitochondrial Bak import and tBid-induced apoptosis. Proc Natl Acad Sci USA 112:E5590–E5599
Gałgańska H, Antoniewicz M, Budzińska M, Gałgański L, Kmita H (2010) VDAC contributes to mRNA levels in Saccharomyces cerevisiae cells by the intracellular reduction/oxidation state dependent and independent mechanisms. J Bioenerg Biomembr 42:483–489
Acknowledgements
VDP acknowledges the financial support by MIUR (PRIN 2015795S5W_005) and University of Catania (Piano Triennale Ricerca). MV acknowledges the financial support by Cariplo Foundation 2015-0641. The authors acknowledge the support of Dr. Giulia Gentile for the initial experimental design of microarray analysis.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Magrì, A., Di Rosa, M., Orlandi, I. et al. Deletion of Voltage-Dependent Anion Channel 1 knocks mitochondria down triggering metabolic rewiring in yeast. Cell. Mol. Life Sci. 77, 3195–3213 (2020). https://doi.org/10.1007/s00018-019-03342-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03342-8