Abstract
Programmable DNA nucleases such as TALENs and CRISPR/Cas9 are emerging as powerful tools for genome editing. Dual-fluorescent surrogate systems have been demonstrated by several studies to recapitulate DNA nuclease activity and enrich for genetically edited cells. In this study, we created a single-strand annealing-directed, dual-fluorescent surrogate reporter system, referred to as C-Check. We opted for the Golden Gate Cloning strategy to simplify C-Check construction. To demonstrate the utility of the C-Check system, we used the C-Check in combination with TALENs or CRISPR/Cas9 in different scenarios of gene editing experiments. First, we disrupted the endogenous pIAPP gene (3.0 % efficiency) by C-Check-validated TALENs in primary porcine fibroblasts (PPFs). Next, we achieved gene-editing efficiencies of 9.0–20.3 and 4.9 % when performing single- and double-gene targeting (MAPT and SORL1), respectively, in PPFs using C-Check-validated CRISPR/Cas9 vectors. Third, fluorescent tagging of endogenous genes (MYH6 and COL2A1, up to 10.0 % frequency) was achieved in human fibroblasts with C-Check-validated CRISPR/Cas9 vectors. We further demonstrated that the C-Check system could be applied to enrich for IGF1R null HEK293T cells and CBX5 null MCF-7 cells with frequencies of nearly 100.0 and 86.9 %, respectively. Most importantly, we further showed that the C-Check system is compatible with multiplexing and for studying CRISPR/Cas9 sgRNA specificity. The C-Check system may serve as an alternative dual-fluorescent surrogate tool for measuring DNA nuclease activity and enrichment of gene-edited cells, and may thereby aid in streamlining programmable DNA nuclease-mediated genome editing and biological research.







Similar content being viewed by others
References
Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF 3rd (2012) Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9:805–807
Kim YG, Chandrasegaran S (1994) Chimeric restriction endonuclease. Proc Natl Acad Sci USA 91:883–887
Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B et al (2009) Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27:851–857
Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC et al (2011) Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29:731–734
Bogdanove AJ, Voytas DF (2011) TAL effectors: customizable proteins for DNA targeting. Science 333:1843–1846
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512
Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ et al (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148
Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, Tan W, Penheiter SG, Ma AC, Leung AY et al (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491:114–118
Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L et al (2011) Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29:695–696
Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 109:17382–17387
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30:460–465
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. eLife 2:e00471
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D et al (2014) Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15:643–652
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M et al (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Shao Y, Guan Y, Wang L, Qiu Z, Liu M, Chen Y, Wu L, Li Y, Ma X, Liu M, Li D (2014) CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc 9:2493–2512
Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, Hotaling JM, Ober C, Hamra FK (2015) Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep 10:1828–1835
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Yosef I, Manor M, Kiro R, Qimron U (2015) Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA 112:7267–7272
Dickinson DJ, Ward JD, Reiner DJ, Goldstein B (2013) Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10:1028–1034
Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110:13904–13909
Hai T, Teng F, Guo R, Li W, Zhou Q (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24:372–375
Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S et al (2015) Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72:1175–1184
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W et al (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843
Wakayama S, Kohda T, Obokata H, Tokoro M, Li C, Terashita Y, Mizutani E, Nguyen VT, Kishigami S, Ishino F, Wakayama T (2013) Successful serial recloning in the mouse over multiple generations. Cell Stem Cell 12:293–297
Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K (2015) Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep 11:875–883
Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E et al (2013) A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31:251–258
Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y, Carlson DF, Bradley P, Bogdanove AJ, Voytas DF (2012) Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PLoS One 7:e45383
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232
Duda K, Lonowski LA, Kofoed-Nielsen M, Ibarra A, Delay CM, Kang Q, Yang Z, Pruett-Miller SM, Bennett EP, Wandall HH et al (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res 42:e84
Feng Y, Zhang S, Huang X (2014) A robust TALENs system for highly efficient mammalian genome editing. Sci Rep 4:3632
Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL et al (2008) Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26:808–816
Ren C, Xu K, Liu Z, Shen J, Han F, Chen Z, Zhang Z (2015) Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell Mol Life Sci 72:2763–2772
Ramakrishna S, Cho SW, Kim S, Song M, Gopalappa R, Kim JS, Kim H (2014) Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun 5:3378
Lim S, Wang Y, Yu X, Huang Y, Featherstone MS, Sampath K (2013) A simple strategy for heritable chromosomal deletions in zebrafish via the combinatorial action of targeting nucleases. Genome Biol 14:R69
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211
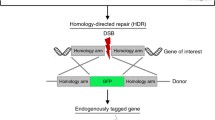
Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113
Kim H, Um E, Cho SR, Jung C, Kim H, Kim JS (2011) Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8:941–943
Liu Y, Lv X, Tan R, Liu T, Chen T, Li M, Liu Y, Nie F, Wang X, Zhou P et al (2014) A modified TALEN-based strategy for rapidly and efficiently generating knockout mice for kidney development studies. PLoS One 9:e84893
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82
Luo Y, Lin L, Bolund L, Sorensen CB (2014) Efficient construction of rAAV-based gene targeting vectors by Golden Gate cloning. Biotechniques 56:263–268
Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T (2007) Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25:786–793
Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, Yamamoto T (2010) Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15:875–885
Kim HK, Kaang BK (1998) Truncated green fluorescent protein mutants and their expression in Aplysia neurons. Brain Res Bull 47:35–41
Luo Y, Lin L, Bolund L, Jensen TG, Sorensen CB (2012) Genetically modified pigs for biomedical research. J Inherit Metab Dis 35:695–713
Holm IE, Alstrup AK, Luo Y (2015) Genetically modified pig models for neurodegenerative disorders. J Pathol. doi:10.1002/path.4654
Zhang X, Cheng B, Gong H, Li C, Chen H, Zheng L, Huang K (2011) Porcine islet amyloid polypeptide fragments are refractory to amyloid formation. FEBS Lett 585:71–77
Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW (2013) Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31:23–24
Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y et al (2013) Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS One 8:e84250
Blechingberg J, Luo Y, Bolund L, Damgaard CK, Nielsen AL (2012) Gene expression responses to FUS, EWS, and TAF15 reduction and stress granule sequestration analyses identifies FET-protein non-redundant functions. PLoS One 7:e46251
Sigal A, Danon T, Cohen A, Milo R, Geva-Zatorsky N, Lustig G, Liron Y, Alon U, Perzov N (2007) Generation of a fluorescently labeled endogenous protein library in living human cells. Nat Protoc 2:1515–1527
Zhou D, Ren JX, Ryan TM, Higgins NP, Townes TM (2004) Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res 32:e128
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542
Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196:961–971
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA 111:E2967–E2976
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3(REVIEWS000):4
Zou L, Luo Y, Chen M, Wang G, Ding M, Petersen CC, Kang R, Dagnaes-Hansen F, Zeng Y, Lv N et al (2013) A simple method for deriving functional MSCs and applied for osteogenesis in 3D scaffolds. Sci Rep 3:2243
Siddle K (2011) Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol 47:R1–R10
Brandl C, Ortiz O, Rottig B, Wefers B, Wurst W, Kuhn R (2015) Creation of targeted genomic deletions using TALEN or CRISPR/Cas nuclease pairs in one-cell mouse embryos. FEBS Open Bio 5:26–35
Levenson AS, Jordan VC (1997) MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res 57:3071–3078
Norwood LE, Grade SK, Cryderman DE, Hines KA, Furiasse N, Toro R, Li Y, Dhasarathy A, Kladde MP, Hendrix MJ et al (2004) Conserved properties of HP1(Hsalpha). Gene 336:37–46
Kirschmann DA, Lininger RA, Gardner LM, Seftor EA, Odero VA, Ainsztein AM, Earnshaw WC, Wallrath LL, Hendrix MJ (2000) Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res 60:3359–3363
Ceccaldi R, Rondinelli B, D’Andrea AD (2015) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol S0962–8924(15):00142–00147
Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30:1473–1475
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:W401–W407
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS (2012) Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22:1327–1333
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Li Y, Park AI, Mou H, Colpan C, Bizhanova A, Akama-Garren E, Joshi N, Hendrickson EA, Feldser D, Yin H et al (2015) A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol 16:111
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147
Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC (2014) Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11:399–402
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32:569–576
Sung P, Klein H (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7:739–750
Voit RA, Hendel A, Pruett-Miller SM, Porteus MH (2014) Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res 42:1365–1378
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Lorson MA, Spate LD, Samuel MS, Murphy CN, Lorson CL, Prather RS, Wells KD (2011) Disruption of the survival motor neuron (SMN) gene in pigs using ssDNA. Transgenic Res 20:1293–1304
Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN et al (2002) Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295:1089–1092
Williams SH, Sahota V, Palmai-Pallag T, Tebbutt SJ, Walker J, Harris A (2003) Evaluation of gene targeting by homologous recombination in ovine somatic cells. Mol Reprod Dev 66:115–125
Du Y, Kragh PM, Zhang Y, Li J, Schmidt M, Bogh IB, Zhang X, Purup S, Jorgensen AL, Pedersen AM et al (2007) Piglets born from handmade cloning, an innovative cloning method without micromanipulation. Theriogenology 68:1104–1110
Luo Y, Lin L, Golas M, Sørensen CB, Bolund L (2015) Targeted porcine genome engineering with TALENs. In: Li X-Q, Jensen TG (eds) Somatic genome manipulation: advances, methods and applications, 1st edn. Springer, Berlin
Luo Y, Bolund L, Sorensen CB (2012) Pig gene knockout by rAAV-mediated homologous recombination: comparison of BRCA1 gene knockout efficiency in Yucatan and Gottingen fibroblasts with slightly different target sequences. Transgenic Res 21:671–676
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Acknowledgments
We are grateful to the FACS CORE facility (with special thanks to Charlotte Christie Petersen) at the Department of Biomedicine, Aarhus University for assistance with flow cytometry and FACS. This work was supported in part by grants from the STAR programme from the R&D Department, Novo Nordisk A/S to YL; and the Danish Research Council for Independent Research (16942) to YL; the Sapere Aude Young Research Talent prize to YL (18382); the Lundbeck Foundation (R173-2014-1105, R151-2013-14439, R126-2012-12448, R100-A9209, R173-2014-993, and R100-A9606) to YL, LB, PB, CBS, and ALN respectively; the China Scholarship Council (CSC) to YZ; the Natural Science Foundation of China (81472126) to ST and GQZ; the Toyota Foundation ALN; and the AUFF AU IDEAS Programme and The Karen Elise Jensen Foundation to CBS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
YL (2012–2014), SGR, and HD were financed by Novo Nordisk A/S. TK was financed by Gubra ApS. A patent claim is declared to the generation of a diabetes pig model based on genetic modification of the porcine IAPP gene. No other competing interests are declared by the authors.
Additional information
Y. Zhou, Y. Liu, D. Hussmann, P. Brøgger contributed equally to the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2015_2128_MOESM3_ESM.pdf
Additional File 3: Optimization of nucleofection in porcine fibroblasts. Five nucleofection reagents (P1-P5) and 15 nucleofection programs were evaluated (PDF 608 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Liu, Y., Hussmann, D. et al. Enhanced genome editing in mammalian cells with a modified dual-fluorescent surrogate system. Cell. Mol. Life Sci. 73, 2543–2563 (2016). https://doi.org/10.1007/s00018-015-2128-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-015-2128-3