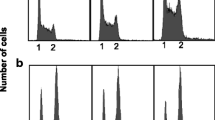
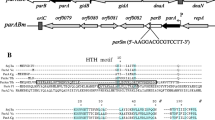
Summary
We have shown that the plasmid pSC101 is unable to be maintained in strains of E. coli carrying deletions in the genes himA and hip which specify the pleitropic heterodimeric DNA binding protein, IHF. We show that this effect is not due to a modulation of the expression of the pSC101 RepA protein, required for replication of the plasmid. Inspection of the DNA sequence of the essential replication region of pSC101 reveals the presence of a site, located between the DnaA binding-site and that of RepA, which shows extensive homology with the consensus IHF binding site. The proximity of the sites suggests that these three proteins, IHF, DnaA, and RepA may interact in generating a specific DNA structure required for initiation of pSC101 replication.
Similar content being viewed by others
References
Bear SE, Court DL, Friedmann DI (1984) An accessory role for E. coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol 52:966–972
Caro L, Berg CM (1971) P1 transduction. In: Methods in enzymology, vol XXI. Academic Press, New York pp 444–458
Casadaban MJ, Chou J, Cohen SN (1980) In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino terminal fragments of exogenous proteins: E. coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol 143:971–980
Chandler M, Pritchard RH (1975) The effect of gene concentration and relative gene dosage on gene output in E. coli. Mol Gen Genet 138:127–141
Chandler M, Galas DJ (1983) Cointegrate formation mediated by Tn9: II. activity of IS1 is modulated by external DNA sequences. J Mol Biol 170:61–91
Churchward G, Linder P, Caro L (1983) The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucl Acids Res 11:5645–5659
Churchward G, Belin D, Nagamine Y (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165–171
Cohen SN, Chang ACY (1977) Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol 132:734–737
Frey J, Chandler M, Caro L (1979) The effects of an E. coli dnaA1s mutation on the plasmids colE1, pSC101, R100.1, and RTF-Tc. Mol Gen Genet 117–126
Friden P, Voelkel K, Sternglanz R, Freundlich M (1984) Reduced expression of the isoleucine and valine enzymes in integration host factor mutants of E. coli. J Mol Biol 172:573–579
Friedman DJ, Olson EJ, Carver D, Gellert M (1984) Synergistic effect of himA and gyrB mutations: evidence that Him functions control expression of ilv and xyl genes. J Bacteriol 157:484–489
Fuller RR, Funnell BE, Kornberg A (1984) The dnaA protein complex with the E. coli chromosome replication origin (oriC) and other DNA sites. Cell 38:889–900
Galas DJ, Schmitz A (1978) DNase footprinting: a simple method for the detection of protein-DNA binding specificty. Nucl Acids Res 5:3157–3170
Gamas P, Galas D, Chandler M (1985) DNA sequence at the end of IS1 required for transposition. Nature 317:458–460
Goosen N, van de Putte P (1984) Regulation of Mu transposition: I. localization of the presumed recognition sites for HimD and ner functions controlling bacteriophage Mu transcription. Gene 30:41–46
Hashimoto-Gotoh T, Sekiguchi M (1977) Mutations to temperature sensitivity in R-plasmid pSC101. J Bacteriol 131:404–412
Hasunuma K, Sekiguchi M (1977) Replication of pSC101 in E. coli K12: requirement for dnaA function. Mol Gen Genet 154:225–230
Hoyt MA, Knight DM, Das A, Miller HI, Echols H (1982) Control of phage λ development by stability and synthesis of cII protein: role of the viral cIII and host hfl, himA and himD genes. Cell 31:565–573
Leong JM, Nunes-Duby S, Lesser CF, Youderian P, Susskind MM, Landy A (1985) The ϕ80 and P22 attachment sites: primary structure and interaction with E. coli integration host factor. J Biol Chem 260:4468–4477
Linder P, Churchward G, Caro L (1983) Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol 170:287–303
Linder P, Churchward G, Guixian X, Yi-Yi Y, Caro L (1985) An essential replication gene, repA, of plasmid pSC101 is autoregulated. J Mol Biol 181:383–393
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor laboratory, Cold Spring Harbor, New York
Miller HI (1981) Multilevel regulation of bacteriophage λ lysogeny by the E. coli himA gene. Cell 25:269–276
Miller HI (1984) Primary structure of the himA gene of E. coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harbor Symp Quant Biol 49:691–698
Miller HI, Kirk M, Echols H (1981) SOS induction and autoregulation of the himA gene for site-specific recombination in E. coli. Proc Natl Acad Sci USA 78:6754–6758
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Nash HA, Mizuuchi K, Weisberg R, Kikuchi Y, Gellert M (1977) Integrative recombination of bacteriophage λ—The biochemical approach to DNA insertions. In: A. Bukhari (ed) DNA insertion elements, plasmids, and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, p 363
Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313
Ross W, Landy A, Kikuchi Y, Nash H (1979) Interaction of int protein with specific sites on λ att DNA. Cell 18:297–307
Rouviere-Yaniv J, Yaniv M, Germond J-E (1979) E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell 17:265–274
Vocke C, Bastia D (1983) DNA-Protein interaction at the origin of DNA replication of the plasmid pSC101. Cell 35:495–502
Weisberg R, Landy A (1983) Site specific recombination in phage lambda. In: Hendrix (ed) Lambda II. Cold Spring Harbor Laboratory. Cold Spring Harbor, New York, pp 211–250
Willis DK, Uhlin BE, Amini KS, Clark AJ (1981) Physical mapping of the srl recA region of E. coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet 183:497–504
Zahn K, Blattner FR (1985) Sequence-induced curvature at the bacteriophage λ origin of replication. Nature 317:451–453
Author information
Authors and Affiliations
Additional information
Communicated by R. Devoret
Rights and permissions
About this article
Cite this article
Gamas, P., Burger, A.C., Churchward, G. et al. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Molec Gen Genet 204, 85–89 (1986). https://doi.org/10.1007/BF00330192
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00330192