Summary
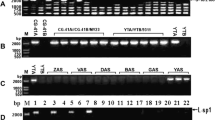
Cloned pearl millet [Pennisetum glaucum (L.) R. Br.] mitochondrial (mt) DNA fragments rearranged by spontaneous reversion from cytoplasmic male sterility (cms) to fertility were characterized by restriction mapping, hybridization with maize mt genes, and transcription analyses. The clones characterized were a 4.7-kb fragment found only in the male-sterile cytoplasm and lost upon reversion to fertility, a 10.9-kb fragment found in all cytoplasms and not changed by reversion, a 13.6-kb fragment found in the male-sterile and -fertile normal cytoplasms and lost in seven of the eight revertants studied, and a 9.7-kb fragment not found in the male-sterile cytoplasm but produced by reversion from male sterility to fertility. The restriction maps verified that the four cloned pearl millet fragments contained two sets of repeated sequences, one on the 4.7-, 10.9-, and 13.6-kb fragments, the other on the 13.6- and 9.7-kb fragments. The rrn18, rrn5, and coxI genes were located in the repeated regions of the 4.7-, 10.9-, and 13.6-kb cloned fragments. The correlation of reversion (eight independent events) with the loss of fragments containing the rrn18, rrn5, and coxI genes suggests that those lost fragments and their gene content could be responsible for the expression of cms. Transcriptional analyses using both Northern blots and end-labeled mtRNA probes verified that transcripts homologous to the rrn18 and coxI genes were present in pearl millet total mtRNA. However, no transcript differences were detected among cms, revertant, and fertile normal cytoplasms, suggesting that the reversion process involves mutational changes that may not affect transcript size. Transcript analyses indicated that the 10.9-kb clone contained an unidentified gene on the end opposite the rrn18 gene; however, since it was present in all cytoplasms, it is not believed to be involved in cms.
Similar content being viewed by others
References
Braun CJ, Levings CS III (1985) Nucleotide sequence of the F1-ATPase alpha subunit gene from maize mitochondria. Plant Physiol 79:571–577
Chao S, Sederoff RR, Levings CS III (1983) Partial sequence analysis of the 5S and 18S rRNA region of the maize mitochondrial genome. Plant Physiol 71:190–193
Chao S, Sederoff RR, Levings CS III (1984) Nucleotide sequence and evolution of the 18S ribosomal RNA gene in maize mitochondria. Nucleic Acids Res 12:6629–6644
Chase CD, Pring DR (1986) Properties of the linear N1 and N2 plasmid-like DNAs from mitochondria of cytoplasm male-sterile Sorghum bicolor. Plant Mol Biol 6:53–64
Dale RMK, Mendu N, Ginsburg H, Kridl JC (1984) Sequence analysis of the maize mitochondrial 26S rRNA gene and flanking regions. Plasmid 11:141–150
Dewey RE, Levings CS III, Timothy DH (1985a) Nucleotide sequence of ATPase subunit 6 gene of maize mitochondria. Plant Physiol 79:914–919
Dewey RE, Schuster AM, Levings CS III, Timothy DH (1985b) Nucleotide sequence of F0-ATPase proteolipid (subunit 9) gene of maize mitochondria. Proc Natl Acad Sci USA 82:1015–1019
Dewey RE, Levings CS III, Timothy DH (1986) Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell 44:439–449
Dewey RE, Timothy DH, Levings CS III (1987) A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc Natl Acad Sci USA 84:5374–5378
Dewey RE, Siedow JN, Timothy DH, Levings CS III (1988) A 13-kilodalton maize mitochondrial protein in E. coli confers sensitivity to Bipolaris maydis toxin. Science 239:293–295
Fauron CMR, Havlik M, Brettell RIS (1990) The mitochondrial genome organization of a maize fertile cms-T revertant line is generated through recombination between two sets of repeats. Genetics 124:423–428
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 137:266–267
Forde BG, Leaver CJ (1980) Nuclear and cytoplasmic genes controlling synthesis of variant mitochondrial polypeptides in male-sterile maize. Proc Natl Acad Sci USA 77:418–422
Fox TD, Leaver CJ (1981) The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell 26:315–323
Hanson MR, Conde MF (1985) Functioning and variation of cytoplasmic genomes: lessons from cytoplasmic-nuclear interactions conferring male sterilities in plants. Int Rev Cytol 94:213–267
Holmes DS, Quigley M (1981) A rapid boiling method for preparation of bacterial plasmids. Anal Biochem 114:193–197
Isaac PG, Jones VP, Leaver CJ (1985) The maize cytochrome c oxidase subunit I gene: sequence expression and rearrangement in cytoplasmic male plants. EMBO J 4:1617–1623
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor /NY
Pearson OH (1981) Nature and mechanisms of cytoplasmic male sterility in plants: a review. Hort Science 16:482–487
Schaffer HE, Sederoff RR (1981) Improved estimation of DNA fragment lengths from agarose gels. Anal Biochem 115:113–122
Smith RL, Chowdhury MKU (1989) Mitochondrial DNA polymorphism in male-sterile and -fertile cytoplasms of pearl millet. Crop Sci 29:809–814
Smith RL, Chowdhury MKU, Pring DR (1987) Mitochondrial DNA rearrangements in Pennisetum associated with reversion from cytoplasmic male sterility to fertility. Plant Mol Biol 9:277–286
Wise RP, Pring DR, Gengenbach BG (1987) Mutation to male fertility and toxin insensitivity in Texas (T)-cytoplasm in maize is associated with a frame shift in a mitochondrial open reading frame. Proc Natl Acad Sci USA 84:2858–2862
Young EG, Hanson M (1987) A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated. Cell 50:41–49
Author information
Authors and Affiliations
Additional information
Communicated by P. L. Pfahler
Florida Agricultural Experiment Station Journal Series No. R-00035
Rights and permissions
About this article
Cite this article
Smith, R.L., Chowdhury, M.K.U. Characterization of pearl millet mitochondrial DNA fragments rearranged by reversion from cytoplasmic male sterility to fertility. Theoret. Appl. Genetics 81, 793–799 (1991). https://doi.org/10.1007/BF00224992
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00224992