Abstract
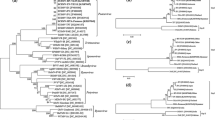
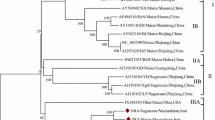
Sunflower chlorotic mottle virus (SuCMoV), the most prevalent virus of sunflower in Argentina, was reported naturally infecting not only sunflower but also weeds. To understand SuCMoV evolution and improve the knowledge on its variability, the complete genomic sequences of two SuCMoV isolates collected from Dipsacus fullonum (-dip) and Ibicella lutea (-ibi) were determined from three overlapping cDNA clones and subjected to phylogenetic and recombination analyses. SuCMoV-dip and -ibi genomes were 9,953-nucleotides (nt) long; their sequences contained an open reading frame of 9,561 nucleotides, which encoded a polyprotein of 3,187 amino acids flanked by a 5′-noncoding region (NCR) of 135 nt and a 3′-NCR of 257 nt. SuCMoV-dip and -ibi genome nucleotide sequences were 90.9 identical and displayed 90 and 94.6 % identity to that of SuCMoV-C, and 90.8 and 91.4 % identity to that of SuCMoV-CRS, respectively. P1 of SuCMoV-dip and -ibi was 3-nt longer than that of SuCMoV-CRS, but 12-nt shorter than that of SuCMoV-C. Two recombination events were detected in SuCMoV genome and the analysis of dN/dS ratio among SuCMoV complete sequences showed that the genomic regions are under different evolutionary constraints, suggesting that SuCMoV evolution would be conservative. Our findings provide evidence that mutation and recombination would have played important roles in the evolutionary history of SuCMoV.


Similar content being viewed by others
References
G. Dujovny, T. Usugi, K. Shohara, S. Lenardon, Plant Dis. 82, 470–474 (1998)
S. Lenardon, in Enfermedades del girasol en la Argentina, manual de reconocimiento, ed. by V. Pereyra. (INTA Centro Regional Balcarce, 1994), pp. 99–103
M.J. Adams, F.M. Zerbini, R. French, F. Rabenstein, D.C. Stenger, J.P.T. Valkonen, in Virus Taxonomy: 9th Report of the International Committee on the Taxonomy of Viruses, ed. by A.M.Q. King, M.J. Adams, E.B. Carstens (Elsevier Academic Press, San Diego, 2011), pp. 1069–1089
A.K. Inoue-Nagata, P.A. Oliveira, L.S. Dutra, T. Nagata, Virus Genes 33, 45–49 (2006)
G. Dujovny, T. Sasaya, H. Koganezawa, T. Usugi, K. Shohara, S. Lenardon, Arch. Virol. 145, 2249–2258 (2000)
F. Giolitti, N. Bejerman, S. de Breuil, S. Lenardon, J. Phytopathol. 158, 536–541 (2010)
N. Bejerman, F. Giolitti, S. de Breuil, S. Lenardon, Arch. Virol. 155, 1331–1335 (2010)
F. Giolitti, N. Bejerman, S. Lenardon, J. Phytopathol 157, 325–328 (2009)
N. Bejerman, F. Giolitti, S. de Breuil, S. Lenardon . (2011). In Book of abstracts, 2nd Phytopathology Argentinean Congress(p. 248). Argentina: Mar del Plata
K. Ohshima, Y. Yamaguchi, R. Hirota, T. Hamamoto, K. Tomimura, Z. Tan, T. Sano, F. Azuhata, J.A. Walsh, J. Fletcher, J. Chen, A. Gera, A. Gibbs, J. Gen. Virol. 83, 1511–1521 (2002)
I.M. Moreno, J.M. Malpica, J.A. Diaz-Pendon, E. Moriones, A. Fraile, F. García-Arenal, Virology 318, 451–460 (2004)
F. García-Arenal, A. Fraile, J.M. Malpica, Annu. Rev. Phytopathol. 39, 157–186 (2001)
T. Ogawa, Y. Tomitaka, A. Nakagawa, K. Ohshima, Virus Res. 131, 199–212 (2008)
A. Padhi, K. Ramu, Virus Genes 42, 282–285 (2011)
S. Wylie, R. Jones, Phytopathology 99, 512–518 (2009)
T. Tsuneyoshi, T. Matsumi, T.C. Deng, I. Sako, S. Sumi, Arch. Virol. 143, 1093–1107 (1998)
D. Huson, D. Bryant, Mol. Biol. Evol. 23, 254–267 (2006)
D.P. Martin, P. Lemey, M. Lott, V. Moulton V. D. Posada, P. Lefeuvre, Bioinformatics 26, 2462–2463 (2010)
K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei, S. Kumar, Mol. Biol. Evol. 28, 2731–2739 (2011)
P. Pamilo, N.O. Bianchi, Mol. Biol. Evol. 10, 271–281 (1993)
W.H. Li, J. Mol. Evol. 36, 96–99 (1993)
S.L. Kosakovsky Pond, S.D.W. Frost, Bioinformatics 21, 2531–2533 (2005)
B. Chung, W.A. Miller, J.F. Atkins, A.E. Firth, Proc. Natl. Acad. Sci. USA 105, 5897–5902 (2008)
X. Xia, Z. Xie, J. Heredity 92, 371–373 (2001)
X. Gu, W. Li, J. Mol. Evol. 40, 464–473 (1995)
J.J. Valli, J.A. Lopez-Moya, J. García, Gen. Virol. 88, 1016–1028 (2007)
R. Krause-Sakate, H. Fakhfakh, M. Peypelut, M. Pavan, F. Zerbini, M. Marrakchi, T. Candresse, O. Le Gall, Arch. Virol. 149, 191–197 (2004)
M. Glasa, T. Malinowski, L. Predajňa, G.N. Pupola, D. Dekena, L. Michalczuk, T. Candresse, Phytopathology 101, 980–985 (2011)
J.K. Seo, K. Ohshima, H.G. Lee, M. Son, H.S. Choi, S.H. Lee, S.H. Sohn, K.H. Kim, Virology 393, 91–103 (2009)
C. Desbiez, H. Lecoq, Arch. Virol. 153, 1749–1754 (2008)
L. Glais, M. Tribodet, C. Kerlan, Arch. Virol. 147, 363–378 (2002)
H.L. Zaaijer, F.J. van Hemert, M.H. Koppelman, V.V. Lukashov, J. Gen. Virol. 88, 2137–2143 (2007)
H.Y. Wang, J.L. Liu, R. Gao, J. Chen, H.Y. Shao, X.D. Li, Virus Genes 38, 421–428 (2009)
B. Moury, C. Morel, E. Johansen, M. Jacquemond, J. Gen. Virol. 83, 2563–2573 (2002)
B. Moury, C. Morel, E. Johansen, S. Souche, V. Ayme, C. Caranta, A. Palloix, M. Jacquemond, Mol. Plant Microbe Interact. 17, 322–329 (2004)
Acknowledgments
We thank INTA for financial support. We are grateful to Dr. E. Taleisnik for editing the manuscript and to Dr. K. Ohshima and Dr. M. Adams for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bejerman, N., Giolitti, F., de Breuil, S. et al. Sequencing of two Sunflower chlorotic mottle virus isolates obtained from different natural hosts shed light on its evolutionary history. Virus Genes 46, 105–110 (2013). https://doi.org/10.1007/s11262-012-0817-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0817-7