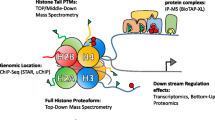
Abstract
Chromatin is a nucleoprotein complex composed of DNA and histone proteins. The concerted activity of chromatin-associated proteins, histone post-translational modifications, and DNA methylation induces epigenetic variations that regulate most of the physiological processes of eukaryotic cells, ranging from gene expression to DNA replication and repair. Epigenetics has also been shown to be tightly linked to cell metabolism. For instance, histone modifications are highly sensitive to the changes in the microenvironment and the local concentration of specific metabolites. Mass-spectrometry (MS)-based proteomics significantly contributed to the recent advances in the epigenetic field, by allowing the comprehensive analysis of histone post-translational modifications as well as the systematic identification of chromatin constituents.
In this chapter, we will provide a general overview of various MS-based experimental strategies developed to boost the epigenetic field, with references to the studies whereby chromatin biology was assessed in relation to cell metabolism.
Similar content being viewed by others
Abbreviations
- AF-10:
-
Antisecretor factor 10
- ChIP:
-
Chromatin immunoprecipitation
- CID:
-
Collision-induced dissociation
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- DDA:
-
Data-dependent acquisition
- ECD:
-
Electron capture dissociation
- ESC:
-
Embryonic stem cell
- ETD:
-
Electron transfer dissociation
- HAT:
-
Histone acetyltransferase
- HMCV:
-
Cytomegalovirus
- hmSILAC:
-
Heavy-methyl SILAC
- KAT:
-
Lysine acetyl-transferase
- LC:
-
Liquid chromatography
- LF:
-
Label-free
- MRM:
-
Multiple reaction monitoring
- MS:
-
Mass-spectrometry
- NPC:
-
Neural progenitor cell
- NSC:
-
Neural stem cell
- PTM:
-
Post-translational modification
- RA:
-
Relative abundance
- RP:
-
Reversed-phase
- SAM:
-
S-adenosyl-methionine
- SILAC:
-
Stable isotope labeling with amino acids in cell culture
- SRM:
-
Selected reaction monitoring
- TAL:
-
Transcription activator-like
- TF:
-
Transcription factor
- WCX-HILIC:
-
Weak-cation exchange hydrophilic interaction liquid chromatography
- XIC:
-
MS-extracted ion chromatogram
References
Akhtar A, Becker PB (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in drosophila. Mol Cell 5:367–375
Alabert C et al (2014) Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16:281–293
Alajem A et al (2015) Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep 10:2019–2031
An M et al (2016) The alteration of H4-K16ac and H3-K27met influences the differentiation of neural stem cells. Anal Biochem 509:92–99
Bartke T et al (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143:470–484
Beli P et al (2012) Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46:212–225
Bonaldi T, Imhof A, Regula JT (2004) A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics 4:1382–1396
Byrum SD et al (2012) ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep 2:198–205
Byrum SD, Taverna SD, Tackett AJ (2013) Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res 41:e195
Choudhary C et al (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Cluntun AA et al (2015) The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab 3:10
Curina A et al (2017) High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins. Genes Dev 31:399–412
Darwanto A et al (2010) A modified “cross-talk” between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J Biol Chem 285:21868–21876
Dejardin J, Kingston RE (2009) Purification of proteins associated with specific genomic Loci. Cell 136:175–186
Eberl HC et al (2013) A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 49:368–378
Fan J et al (2015) Metabolic regulation of histone post-translational modifications. ACS Chem Biol 10:95–108
Fraga MF et al (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391–400
Fujita T et al (2013) Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep 3:3171
Gao J et al (2014) Absolute quantification of histone PTM marks by MRM-based LC-MS/MS. Anal Chem 86:9679–9686
Guo A et al (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 13:372–387
Hebert AS et al (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49:186–199
Henry RA et al (2016) Interaction with the DNA repair protein thymine DNA Glycosylase regulates histone acetylation by p300. Biochemistry 55:6766–6775
Islam K et al (2012) Bioorthogonal profiling of protein methylation using azido derivative of S-adenosyl-L-methionine. J Am Chem Soc 134:5909–5915
Jaffe JD et al (2013) Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet 45:1386–1391
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Ji X et al (2015) Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc Natl Acad Sci U S A 112:3841–3846
Jung HR et al (2010) Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9:838–850
Kim J et al (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7:397–403
Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184:868–871
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Kustatscher G et al (2014) Chromatin enrichment for proteomics. Nat Protoc 9:2090–2099
Lambert JP et al (2009) A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics 8:870–882
Lange M et al (2008) Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev 22:2370–2384
Larsen SC et al (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal 9:rs9
Lee JV et al (2014) Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab 20:306–319
Leroy G et al (2013) A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin 6:20
Li X et al (2012) Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc 134:1982–1985
Maile TM et al (2015) Mass spectrometric quantification of histone post-translational modifications by a hybrid chemical labeling method. Mol Cell Proteomics 14:1148–1158
Mews P et al (2014) Histone methylation has dynamics distinct from those of histone acetylation in cell cycle reentry from quiescence. Mol Cell Biol 34:3968–3980
Migliori V et al (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol 19:136–144
Mitchell L et al (2013) mChIP-KAT-MS, a method to map protein interactions and acetylation sites for lysine acetyltransferases. Proc Natl Acad Sci U S A 110:E1641–E1650
Mittler G, Butter F, Mann M (2009) A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res 19:284–293
Mohammed H et al (2016) Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc 11:316–326
Moore KE et al (2013) A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell 50:444–456
Morrish F et al (2010) Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem 285:36267–36274
Nie L et al (2017) The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Mol Cell Proteomics 16:1324–1334
Nikolov M et al (2011) Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol Cell Proteomics M110(005371):10
Noberini R et al (2016) Pathology tissue-quantitative mass spectrometry analysis to profile histone post-translational modification patterns in patient samples. Mol Cell Proteomics 15:866–877
O’Connor CM et al (2014) Quantitative proteomic discovery of dynamic epigenome changes that control human cytomegalovirus (HCMV) infection. Mol Cell Proteomics 13:2399–2410
Ong SE, Mann M (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat Protoc 1:2650–2660
Ong SE, Mittler G, Mann M (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods 1:119–126
Pesavento JJ, Mizzen CA, Kelleher NL (2006) Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem 78:4271–4280
Rafiee MR et al (2016) Expanding the circuitry of Pluripotency by selective isolation of chromatin-associated proteins. Mol Cell 64:624–635
Rardin MJ et al (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A 110:6601–6606
Sansoni V et al (2014) The histone variant H2A.B.bd is enriched at sites of DNA synthesis. Nucleic Acids Res 42:6405–6420
Shiio Y et al (2003) Quantitative proteomic analysis of chromatin-associated factors. J Am Soc Mass Spectrom 14:696–703
Shyh-Chang N et al (2013) Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339:222–226
Sidoli S, Cheng L, Jensen ON (2012) Proteomics in chromatin biology and epigenetics: elucidation of post-translational modifications of histone proteins by mass spectrometry. J Proteome 75:3419–3433
Sidoli S et al (2014) Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics 14:2200–2211
Sirbu BM, Couch FB, Cortez D (2012) Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7:594–605
Soldi M, Bonaldi T (2013) The proteomic investigation of chromatin functional domains reveals novel synergisms among distinct heterochromatin components. Mol Cell Proteomics 12:764–780
Soldi M, Bremang M, Bonaldi T (2014) Biochemical systems approaches for the analysis of histone modification readout. Biochim Biophys Acta 1839:657–668
Spruijt CG et al (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152:1146–1159
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Syka JE et al (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A 101:9528–9533
Tammen SA, Friso S, Choi SW (2013) Epigenetics: the link between nature and nurture. Mol Asp Med 34:753–764
Thomas CE, Kelleher NL, Mizzen CA (2006) Mass spectrometric characterization of human histone H3: a bird’s eye view. J Proteome Res 5:240–247
Torrente MP et al (2011) Proteomic interrogation of human chromatin. PLoS One 6:e24747
Vermeulen M et al (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58–69
Wang CI et al (2013) Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in drosophila. Nat Struct Mol Biol 20:202–209
Wu C, Morris JR (2001) Genes, genetics, and epigenetics: a correspondence. Science 293:1103–1105
Yang YY, Ascano JM, Hang HC (2010) Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc 132:3640–3641
Young NL et al (2009) High throughput characterization of combinatorial histone codes. Mol Cell Proteomics 8:2266–2284
Zee BM, Young NL, Garcia BA (2011) Quantitative proteomic approaches to studying histone modifications. Curr Chem Genomics 5:106–114
Zubarev RA et al (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem 72:563–573
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this entry
Cite this entry
Nicosia, L. et al. (2017). Mass Spectrometry and Epigenetics. In: Patel, V., Preedy, V. (eds) Handbook of Nutrition, Diet, and Epigenetics. Springer, Cham. https://doi.org/10.1007/978-3-319-31143-2_115-1
Download citation
DOI: https://doi.org/10.1007/978-3-319-31143-2_115-1
Received:
Accepted:
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-31143-2
Online ISBN: 978-3-319-31143-2
eBook Packages: Springer Reference MedicineReference Module Medicine