Abstract
Biological databases are growing at an exponential rate, currently being among the major producers of Big Data, almost on par with commercial generators, such as YouTube or Twitter. While traditionally biological databases evolved as independent silos, each purposely built by a different research group in order to answer specific research questions; more recently significant efforts have been made toward integrating these heterogeneous sources into unified data access systems or interoperable systems using the FAIR principles of data sharing. Semantic Web technologies have been key enablers in this process, opening the path for new insights into the unified data, which were not visible at the level of each independent database. In this chapter, we first provide an introduction into two of the most used database models for biological data: relational databases and RDF stores. Next, we discuss ontology-based data integration, which serves to unify and enrich heterogeneous data sources. We present an extensive timeline of milestones in data integration based on Semantic Web technologies in the field of life sciences. Finally, we discuss some of the remaining challenges in making ontology-based data access (OBDA) systems easily accessible to a larger audience. In particular, we introduce natural language search interfaces, which alleviate the need for database users to be familiar with technical query languages. We illustrate the main theoretical concepts of data integration through concrete examples, using two well-known biological databases: a gene expression database, Bgee, and an orthology database, OMA.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Data integration
- Ontology-based data access
- Knowledge representation
- Query processing
- Keyword search
- Relational databases
- RDF stores
1 Introduction
Biological databases have grown exponentially in recent decades, both in number and in size, owing primarily to modern high-throughput sequencing techniques [1]. Today, the field of genomics is almost on par with the major commercial generators of Big Data, such as YouTube or Twitter, with the total amount of genome data doubling approximately every 7 months [2]. While most biological databases have initially evolved as independent silos, each purposely built by a different research group in order to collect data and respond to a specific research question, more recently significant efforts have been made toward integrating the different data sources, with the aim of enabling more powerful insights from the aggregated data, which would not be visible at the level of individual databases.
Let us consider the following example. An evolutionary biologist might want to answer the question “What are the human-rat orthologs, expressed in the liver, that are associated with leukemia?”. Getting an answer for this type of question usually requires information from at least three different sources: an orthology database (e.g., OMA [3], OrthoDB [4], or EggNog [5]); a gene expression database, such as Bgee [6]; and a proteomics database containing disease associations (e.g., UniProt [7]). In the lack of a unified access to the three data sources, obtaining this information is a largely manual and time-consuming process. First, the biologist needs to know which databases to search through. Second, depending on the interface provided by these databases, he or she might need to be familiar with a technical query language, such as SQL or SPARQL (note: a list of acronyms is provided at the beginning of this chapter). At the very least, the biologist is required to know the specific identifiers (IDs) and names used by the research group that created the database, in order to search for relevant entries. An integrated view, however, would allow the user to obtain this information automatically, without knowing any of the details regarding the structure of the underlying data sources—nor the type of storage these databases use—and eventually not even specific IDs (such as protein or gene names).
Biological databases are generally characterized by a large heterogeneity, not only in the type of information they store but also in the model of the underlying data store they use—examples include relational databases, file-based stores, graph based, etc. Examples of databases considered fundamental to research in the life sciences can be found in the ELIXIR Europe’s Core Data Resources, available online at https://www.elixir-europe.org/platforms/data. In this chapter we will mainly discuss two types of database models: the relational model (i.e., relational databases) and a graph-based data model, RDF (the Resource Description Framework).
Database systems have been around since arguably the same time as computers themselves, serving initially as “digitized” copies of tabular paper forms, for example, in the financial sector, or for managing airline reservations. Relational databases, as well as the mathematical formalism underlying them, namely, the relational algebra, were formalized in the 1970s by E.F. Codd, in a foundational paper that now has surpassed 10,000 citations [8]. The relational model is designed to structure data into so-called tuples, according to a predefined schema. Tuples are stored as rows in tables (also called “relations”). Each table usually defines an entity, such as an object, a class, or a concept, whose instances (the tuples) share the same attributes. Examples of relations are “Gene”, “Protein”, “Species”, etc. The attributes of the relation will represent the columns of the table, for example, “gene name.” Furthermore, each row has a unique identifier. The column (or combination of columns) that stores the unique identifier is called a primary key and can be used not only to uniquely identify rows within a table but also to connect data between multiple tables, through a Primary Key-Foreign key relationship. Doing such a connection is called a join. In fact, a join is only one of the operations defined by relational algebra. Other common operations include projection, selection, and others. The operands of relational algebra are the database tables, as well as their attributes, while the operations are expressed through the Structured Query Language (SQL). For a more in-depth discussion on relational algebra, we refer the reader to the original paper by E.F. Codd [8].
This chapter is structured as follows. In Sect. 2, we give a brief introduction to relational databases, through the concrete example of the Bgee gene expression database. We introduce the basics of Semantic Web technologies in Sect. 3. Readers who are already familiar with the Semantic Web stack might skip Sect. 3 and jump directly to Sect. 4, which presents an applied use case of Semantic Web technologies in the life sciences: modeling the Bgee and OMA databases. Section 5 represents the core of this chapter. Here, we present ontology-based data integration (Sect. 5.1) and illustrate it through the concrete example of a unified ontology for Bgee and OMA (Sect. 5.2), as well as the mechanisms required to further extend the integrated system with other heterogeneous sources such as the UniProt protein knowledge base (Sect. 5.3). We introduce natural language interfaces, which enable easy data access even for nontechnical users, in Sect. 5.4. We present an extensive timeline of milestones in data integration based on Semantic Web technologies in the field of life sciences in Sect. 6. Finally, we conclude in Sect. 7.
2 Modeling a Biological Database with Relational Database Technology
In this section we will demonstrate how to model a biological database with relational database technology.
Figure 1 illustrates the data model of a sample extracted from the Bgee database. The sample contains five tables and their relationships, shown as arrows, where the direction of the arrow is oriented from the foreign key of one table to the primary key of a related one. For example, the Primary Key (PK) of the Species table is the SpeciesID. Following the relationships highlighted in bold, we see that the SpeciesID also appears in the two tables connected to Species: GlobalCond and Gene. In these tables, the attribute plays the role of a Foreign Key (FK). The PK-FK relationships allow combining or aggregating data from related tables. For example, by joining Species and Gene, through the SpeciesID, we can find to which species a gene belongs. Concretely, let’s assume we want to find the species where the gene “HBB” can be found. Given that this information is stored in the SpeciesCommonName attribute, we can retrieve it through the following SQL query:
SELECT SpeciesCommonName from Species JOIN Gene WHERE Gene.GeneName = ’HBB’ and Species.SpeciesID = Gene.SpeciesID
This query enables retrieving (via the “SELECT” keyword) the attribute corresponding to the species name (SpeciesCommonName) by joining the Species and Gene tables, based on their primary key-foreign key relationship, namely, via the SpeciesID, on the condition that the GeneName exactly matches “HBB.” For a more detailed introduction to the syntax and usage of SQL, we refer the reader to an online introductory tutorial [9], as well as the more comprehensive textbooks [10, 11].
Taking this a step further, we can imagine the case where a second relational database also stores information about genes, but perhaps with some additional data, such as associations with diseases. Can we still combine information across these distinct databases? Indeed, as long as there is a common point between the tables in the two databases, such as the GeneID or the SpeciesID, it is usually possible to combine them into a single, federated database and use SQL to query it through federated joins. An example of using federated databases for biomedical data is presented in [12].
2.1 Limitations of Relational Databases and Emerging Solutions for Data Integration
So far, we have seen that relational databases are a mature, highly optimized technology for storing and querying structured data. Also, combined with a powerful and expressive query language, SQL, they allow users to federate (join) data even from different databases.
However, there are certain relationships that are not natural for relational databases. Let us consider the relationship “hasOrtholog”. Both the domain and the range of this relationship, as defined in the Orthology Ontology [13], are the same—a gene. For example, the hemoglobin (HBB) gene in human has the Hbb-bt orthologous gene in the mouse (expressed via the relation hasOrtholog). In the relational database world, this translates into a so-called self-join. As the name suggests, this requires joining one table—in this case, Gene—with itself, in order to retrieve the answer. These types of “self-join” relations, while frequent in the real world (e.g., a manager of an employee is also an employee, a friend of a person is also a person, etc.), are inefficient in the context of relational databases. While there are sometimes ways to avoid self-joins, these require even more advanced SQL fluency on the part of the programmer [14].
Moreover, relational databases are typically not well-suited for applications that require frequent schema changes. Hence, NoSQL stores have gained widespread popularity as an alternative to traditional relational database management systems [15,16,17]. These systems do not impose a strict schema on the data and are therefore more flexible than relational databases in the cases where the structure of the data is likely to change over time. In particular, graph databases, such as Virtuoso [18], are very well suited for data integration, as they allow easily combining multiple data sources into a single graph. We discuss this in more detail in Sect. 3.
These and other considerations have led to the vision of the Semantic Web, formalized in 2001 by Tim Berners Lee et al. [19]. At a high-level, the Semantic Web allows representing the semantics of data in a structured, easy to interlink, machine-readable way, typically by use of the Resource Description Framework (RDF)—a graph-based data model. The gradual adoption of RDF stores, although widespread in the Web context and in the life sciences in particular, did not replace relational databases altogether, which lead to a new challenge: how will these heterogeneous data sources now be integrated?
Initial integration approaches in the field of biological databases have been largely manual: first, many of them (either relational or graph-based) have included cross-references to other sources. For example, UniProt contains links to more than 160 other databases. However, this raises a question for the user: which of the provided links should be followed in order to find relevant connections? While a user can be assumed to know the contents of a few related databases, we can hardly expect anyone to be familiar with more than 160 of them! To avoid this problem, other databases have chosen an orthogonal approach: instead of referencing links to other sources, simply copy the relevant data from those sources into the database. This approach also has a few drawbacks. First, it generates redundant data (which might result in significant storage space consumption), and, most importantly, it might lead to the use of stale, outdated results. Moreover, this approach is contradictory to best practices of data warehousing used widely across various domains in industry. For a discussion on this, we refer the reader to [20].
Databases such as UniProt are highly comprehensive, with new results being added to each release, results that may sometimes even contradict previous results. Duplication of this data into another database can quickly lead to missing out the most recent information or to high maintenance efforts required to keep up with the new changes. In the following sections, we discuss an alternative approach: integrating heterogeneous data sources through the use of a unifying data integration layer, namely, an integrative ontology, that aligns, but also enriches the existing data, with the purpose of facilitating knowledge discovery.
Throughout the remainder of this chapter, we will combine theoretical aspects of data integration with concrete examples, based on our SODA project [21], as well as from our ongoing research project, Bio-SODA [22], where we are currently building an integrated data access system for biological databases (starting with OMA and Bgee), using a natural language search interface. In the context of this project, Semantic Web technologies, such as RDF, are used to enhance interoperability among heterogeneous databases at the semantic level (e.g., RDF graphs with predefined semantics). Moreover, currently, several life science and biomedical databases such as OMA [3], UniProt [7], neXtProt [22], the European Bioinformatics Institute (EMBL-EBI) RDF data [24], and the WorldWide Protein Data Bank [25] already provide RDF data access, which also justifies an RDF-based approach to enable further integration efforts to include these databases. A recent initiative for (biological) data sharing is based on the FAIR principles [26], aiming to make data findable, accessible, interoperable, and re-usable.
3 Semantic Web Technologies
The Semantic Web, as its name shows, emerged mainly as a means to attach semantics (meaning) to data on the Web [19]. In contrast to relational databases, Semantic Web technologies rely on a graph data model, in order to enable interlinking data from disparate sources available on the Web. Although the vision of the Semantic Web still remains an ideal, many large datasets are currently published based on the Linked Data principles [27] using Semantic Web technologies (e.g., RDF). The Linked Open Data Cloud illustrates a collection of a large number of different resources including DBPedia, UniProt, and many others.
In this section, we will describe the Semantic Web (SW) stack, focusing on the technologies that enhance data integration and enrichment. For a more complete description of the SW stack, we refer the reader to the comprehensive introductions in [28,29,30].
The Semantic Web stack is presented in Fig. 2. We will focus on the following standards or layers of the stack: URI, the syntax layer (e.g., Turtle (TTL), an RDF serialization format), RDF, OWL, RDFS, and SPARQL. These layers are highlighted in gray in Fig. 2.
The Semantic Web stack modified from [31]
3.1 Unique Resource Identifier (URI)
A Uniform Resource Identifier (URI) is a character sequence that identifies an abstract or physical resource. A URI is classified as a locator, a name, or both. The Uniform Resource Locators (URLs) are a subset of URIs that, in addition to identifying a resource, provide a means of locating the resource by describing its primary access or network “location.” For example, https://bgee.org is a URI that identifies a resource (i.e., the Bgee gene expression website), and it implies solely a representation of this resource (i.e., an HTML Web page). This resource is accessible through the HTTPS protocol.
The Uniform Resource Name (URN) is also a URI that refers to both the “urn” scheme [32], which are URIs required to remain globally unique and persistent even when the resource does not exist anymore or becomes unavailable, and to any other URI with the properties of a name. For example, the URN urn:isbn:978-1-61779-581-7 is a URI that refers to a previous edition of this book by using the International Standard Book Number (ISBN). However, no information about the location and how to get this resource (book) is provided.
The URI syntax consists of a hierarchical sequence of components referred to as the scheme, authority, path, query, and fragment [33]. Figure 3 describes a UniProt URI that includes these components.
An individual scheme does not have to be classified as being just one of “name” or “locator.” Instances of URIs from any given scheme may have the characteristics of names (URN) or locators (URL) or both (URN + URL). Further examples of URIs with variations in their syntax components are:
-
ftp://ftp.bgee.org/current/download/calls/expr_calls/Sus_scrofa_expr_simple_development.tsv.zip
-
mailto:Bgee@sib.swiss
-
urn:miriam:pubmed:26615188
3.2 Resource Description Framework (RDF)
The Resource Description Framework (RDF) is a framework for describing information about resources in the World Wide Web, which are identified with URIs. In the previous section, we have seen that data in relational databases is organized into tables, according to some predefined schema. In contrast, in RDF stores, data is mainly organized into triples, namely, <subject, predicate, object>, similarly to how sentences in natural language are structured. An informal example would be: <Bob, isFriendOf, Alice>. A primer on triples and the RDF data model, using this simple example, is available online [34]. Figure 4 illustrates the RDF triple: the subject represents the resource being described, the predicate is a property of that resource, and finally the object is the value of the property (i.e., an attribute of the subject).
Triples can be defined using the RDF. The data store for RDF data is also called a “triple store.” Moreover, in analogy to the data model (or the schema) of a relational database, the high-level structure of data in a triple store can be described using an ontology. According to Studer et al. [35], an ontology is a formal, explicit specification of a shared conceptualization. “Formal” refers to the fact that the expressions must be machine readable: hence, natural language is excluded. In this context, we can mention description logic (DL)-based languages [36], such as OWL 2 DL (see Sect. 3.3 for further details) to define ontologies. A DL ontology is the equivalent of a knowledge base (KB). A KB is mainly composed of two components that describe different statements in ontologies: the terminological box (TBox, i.e., the schema) and the assertional box (ABox, i.e., the data). Therefore, the conceptual statements form the set of TBox axioms, whereas the instance level statements form the set of ABox assertions. To exemplify this, we can mention the following DL axioms: Man ≡ Human ⊓ Male (a TBox axiom that states a man is a human and male) and john:Man (an ABox assertion that states john is an instance of man).
Given that one of the goals of the Semantic Web is to assign unambiguous names to resources (URIs), an ontology should be more than a simple description of data in a particular triple store. Rather, it should more generally serve as a description of a domain, for instance, genomics (see Gene Ontology [37]) or orthology (see Orth Ontology [13]). Different instantiations of this domain, for example, by different research groups, should reuse and extend this ontology. Therefore, constructing good ontologies requires careful consideration and agreement between domain specialists, with the goal of formally representing knowledge in their field. As a consequence, ontologies are usually defined in the scope of consortiums—such as the Gene Ontology Consortium [38] or the Quest for Orthologs Consortium [39]. A notable collaborative effort is the Open Biological and Biomedical Ontology (OBO) Foundry [40]. It established principles for ontology development and evolution, with the aim of maximizing cross-ontology coordination and interoperability, and provides a repository of life science ontologies, currently, including about 140 ontologies.
To give an example of RDF data in a concrete life sciences use case, let us consider the following RDF triples, which illustrate a few of the assertions used in the OMA orthology database to describe the human hemoglobin protein (“HBB”), using the first version of the ORTH ontology [13]:
oma:PROTEIN_HUMAN04027 rdf:type orth:Protein. oma:PROTEIN_HUMAN04027 oma:geneName “HBB”. oma:PROTEIN_HUMAN04027 biositemap:description “Hemoglobin subunit beta". oma:PROTEIN_HUMAN04027 obo:RO_0002162 <http://www.uniprot.org/taxonomy/9606>.
This simple example already illustrates most of the basics of RDF. The instance that is being defined—the HBB protein in human—has the following URI in the OMA RDF store: http://omabrowser.org/ontology/oma#PROTEIN_HUMAN04027
The URI is composed of the OMA prefix, http://omabrowser.org/ontology/oma# (abbreviated here as “oma:”), and a fragment identifier, PROTEIN_HUMAN04027. The first triple describes the type of this resource—namely, an orth:Protein—based on the Orthology Ontology, prefixed here as “orth:,” http://purl.org/net/orth#. As mentioned previously, this is a higher-level ontology, which OMA reuses and instantiates. It is important to note that other ontologies are used as well in the remaining assertions: for example, the last triple references the UniProt taxonomy ID 9606. This is based on the National Center for Biotechnology Information (NCBI) organismal taxonomy [41]. If we follow the link in a Web browser, we see that it identifies the “Homo sapiens” species, while the property obo:RO_0002162 (i.e., http://purl.obolibrary.org/obo/RO_0002162) simply denotes “in taxon” in OBO [40]. Lastly, the concept also has a human-readable description, “Hemoglobin subunit beta.”
3.3 RDF Schema (RDFS)
RDF Schema (RDFS) provides a vocabulary for modeling RDF data and is a semantic extension of RDF. It provides mechanisms for describing groups (i.e., classes) of related resources and the relationships between these resources. The RDFS is defined in RDF. The RDFS terms are used to define attributes of other resources such as the domains (rdfs:domain) and ranges (rdfs:range) of properties. Moreover, the RDFS core vocabulary is defined in a namespace informally called rdfs here, and it is conventionally associated with the prefix rdfs:. That namespace is identified by the URI http://www.w3.org/2000/01/rdf-schema# .
In this section, we will mostly focus on the RDF and RDFS terms used in this chapter. Further information about RDF/RDFS terms is available in [42].
-
Classes
-
rdfs:Resource—all things described by RDF are called resources, which are instances of the class rdfs:Resource (i.e., rdfs:Resource is an instance of rdfs:Class).
-
rdfs:Class is the class of resources that are RDF classes. Resources that have properties (attributes) in common may be divided into classes. The members of a class are instances.
-
rdf:Property is a relation between subject and object resources, i.e., a predicate. It is the class of RDF properties.
-
rdfs:Literal is the class of literal values such as textual strings and integers. rdfs:Literal is a subclass of rdfs:Resource.
-
-
Properties
-
rdfs:range is an instance of rdf:Property. It is used to state that the values of a property are instances of one or more classes. For example, orth:hasHomolog rdfs:range orth:SequenceUnit (see Fig. 5a). This statement means that the values of orth:hasHomolog property can only be instances of orth:SequenceUnit class.
-
rdfs:domain is an instance of rdf:Property. It is used to state that any resource that has a given property is an instance of one or more classes. For example, orth:hasHomolog rdfs:domain orth:SequenceUnit (see Fig. 5b). This statement means that resources that assert the orth:hasHomolog property must be instances of orth:SequenceUnit class.
-
rdf:type is an rdf:Property that is used to state that a resource is an instance of a class.
-
rdfs:subClassOf is an rdf:Property to assert that all instances of one class are instances of another. For example, if C1 rdfs:subClassOf C2 then an instance of C1 is also an instance of C2 but not vice versa.
-
rdfs:subPropertyOf is used to state that all resources related by one property (i.e., the subject of rdfs:subPropertyOf) are also related by another (i.e., the object of rdfs:subPropertyOf, the “super-property”). For example, all orthologous relations are also homologous relations. Because of this, in the latest release candidate of the Orthology Ontology [13], it is stated that orth:hasOrtholog is a sub-property of orth:hasHomolog. Figure 5c illustrates this statement.
-
3.4 Web Ontology Language (OWL)
The first level above RDF/RDFS in the Semantic Web stack (see Fig. 2) is an ontology language that can formally describe the meaning of resources. If machines are expected to perform useful reasoning tasks on RDF data, the language must go beyond the basic semantics of RDF Schema [43]. Because of this, OWL and OWL 2 (i.e., Web Ontology languages) include more terms for describing properties and classes, such as relations between classes (e.g., disjointness, owl:disjointWith), cardinality (e.g., “exactly 2,” owl:cardinality), equality (i.e., owl:equivalentClass), richer typing of properties, characteristics of properties (e.g., symmetry, owl:SymmetricProperty), and enumerated classes (i.e., owl:oneOf). The owl: prefix replaces the following URI namespace: http://www.w3.org/2002/07/owl#.
As a full description of OWL and OWL 2 is beyond the scope of this chapter, we refer the interested reader to [44, 45]. In the following, we focus solely on some essential modeling features that the OWL languages offer in addition to RDF/RDFS vocabularies.
-
owl:Class is a subclass of rdfs:Class. Like rdfs:Class, an owl:Class groups instances that share common properties. However, this new OWL term is defined due to the restrictions on DL-based OWL languages (e.g., OWL DL and OWL Lite; OWL 2 DL and its syntactic fragments EL, QL, and RL). These restrictions imply that not all RDFS classes are legal OWL DL/OWL 2 DL classes. For example, the orth:SequenceUnit entity in the ORTH ontology is stated as an OWL class (i.e., orth:SequenceUnit rdf:type owl:Class—Fig. 5d illustrates this axiom). Therefore, orth:SequenceUnit is also an RDFS class since owl:Class is a subclass of rdfs:Class.
-
owl:ObjectProperty is a subclass of rdf:Property. The instances of owl:ObjectProperty are object properties that link individuals to individuals (i.e., members of an owl:Class). For example, the orth:hasHomolog object property (see Fig. 5e) relates one orth:SequenceUnit individual to another one. Figure 5a illustrates this example.
-
owl:DatatypeProperty is a subclass of rdf:Property. The instances of owl:DatatypeProperty are datatype properties that link individuals to data values. To illustrate a datatype property, we can mention the oma:ensemblGeneId (see Figs. 5f and 6b). This property asserts a gene identifier to an instance of an orth:Gene.
Further information about OWL languages are available as World Wide Web Consortium (W3C) recommendations in [46] and [47].
3.5 RDF Serialization Formats
RDF is a graph-based data model which provides a grammar for its syntax. Using this grammar, RDF syntax can be written in various concrete formats which are called RDF serialization formats. For example, we can mention the following formats: Turtle [48], RDF/XML (an XML syntax for RDF) [49], and JSON-LD (a JSON syntax for RDF) [50]. In this section, we will solely focus on the Turtle format.
Turtle language (TTL) allows for writing an RDF graph in a compact textual form. To exemplify this serialization format, let us consider the following turtle document that defines the homologous and orthologous relations:
@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> . @prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> . @prefix owl: <http://www.w3.org/2002/07/owl#> . @prefix orth: <http://purl.org/net/orth#> . # http://purl.org/net/orth#SequenceUnit orth:SequenceUnit rdf:type owl:Class . orth:hasHomolog rdf:type owl:ObjectProperty ; rdf:type owl:SymmetricProperty ; rdfs:domain orth:SequenceUnit ; rdfs:range orth:SequenceUnit . orth:hasOrtholog rdf:type owl:ObjectProperty ; rdfs:subPropertyOf orth:hasHomolog .
This example introduces many of features of the Turtle language: @prefix and prefixed names (e.g., @prefix rdfs: http://www.w3.org/2000/01/rdf-schema# ), predicate lists separated by “;” (e.g., orth:hasOrtholog rdf:type owl:ObjectProperty; rdfs:subPropertyOf orth:hasHomolog.), comments prefixed with “#” (e.g., # http://purl.org/net/orth#SequenceUnit), and a simple triple where the subject, predicate, and object are separated by white spaces and ended with a “.” (e.g., orth:SequenceUnit rdf:type owl:Class).
Further details about TTL serialization are available as a W3C recommendation in [48]
3.6 Querying the Semantic Web with SPARQL
Once we have defined the knowledge base (TBox and ABox), how can we use it to retrieve relevant data? Similar to SQL for relational databases, data in RDF stores can be accessed by using a query language. One of the main RDF query languages, especially used in the field of life sciences, is SPARQL [51]. A SPARQL query essentially consists of a graph pattern, namely, conjunctive RDF triples, where the values that should be retrieved (the unknowns—either subjects, predicates, or objects) are replaced by variable names, prefixed by “?”. Looking again at the previous example, if we want to get the description of the “HBB” protein from OMA, we would simply use a graph pattern, where the value of the “description”—the one we want to retrieve—is replaced by a variable as follows:
SELECT ?description WHERE { ?protein oma:geneName “HBB”. ?protein biositemap:description ?description. }
The choice of variable name itself is not important (we could have used “?x”, “?var”, etc., albeit with a loss of readability). Essentially, we are interested in the description of a protein about which we only know a name—“HBB.”
In order to get a sense of how large bioinformatics databases currently are, but also to get a hands-on introduction into how they can be queried using SPARQL, we propose to retrieve the total number of proteins in UniProt in Exercise A at the end of this chapter. Furthermore, Exercise C will allow trying out and refining the OMA query introduced above, but also writing a new one, using the OMA SPARQL endpoint.
4 Modeling Biological Databases with Semantic Web Technologies
In this section we show a concrete example of how we can use Semantic Web technologies to model the two biology databases Bgee and OMA.
Figure 7 illustrates a fragment of a candidate ontology describing the relational database sample from Bgee (see Fig. 1). The ellipses illustrate classes of the ontology, either specific to the Bgee ontology, such as AnatomicEntity (the equivalent of the anatEntity table in the relational view), or classes from imported ontologies, such as the Taxon class (the prefix “up:” denoting the UniProt ontology, http://purl.uniprot.org/core/). The advantage of using external (i.e., imported) classes is that integration with other databases which also instantiate these classes will be much simpler. For example, we will see that the class Gene serves as the “join point” between OMA and Bgee. Arrows define properties of the ontology: either datatype properties (similar to attributes of a table in the relational world), such as the speciesName or the stageName, or object properties, which are similar to primary key-foreign key relationships, given that they link instances of one class to those of another. If we compare Fig. 7 (the ontology view) against Fig. 1 (the relational view), we notice that the object properties isExpressedIn and isAbsentIn only appear explicitly in the ontology. This is because the values of these properties will actually be calculated on-the-fly, from multiple attributes in the relational database. Given that Bgee is mainly used to query gene expressions, these properties are exposed as new semantic properties in the domain ontology, namely, expression or absence of expression of a gene in a particular anatomic entity. This is one of the means through which the semantic layer can not only describe but also enrich the data available in the underlying layers (in this case, in the relational database). The domain of both the isExpressedIn and isAbsentIn properties is in this case a gene, while the range is an anatomic entity, such that triples that instantiate this relationship will have the structure: <Gene, isExpressedIn, AnatomicEntity>.
Given that the OMA ontology is significantly larger than the one for Bgee, we only show here the class hierarchy in Fig. 8. The most important concepts in the ontology are shown in the top right corner, namely, the cluster of orthologs and the cluster of paralogs, which store information about gene orthology (or paralogy) in a hierarchical tree structure (the gene-tree node). Similarly to the Bgee ontology, the Gene class in OMA is external. Arrows indicate the “rdfs:subClassOf” relationship—for example, both the “Cluster of Orthologs” and the “Cluster of Paralogs” classes—are subclasses of the “Cluster of Homologs” class. For a description of the ontology, as well as a discussion regarding its design within the Quest for Orthologs Consortium, we point the reader to [13]. Furthermore, the ontology can be explored or visualized in WebVOWL [52] using the Web page of the OMA SPARQL endpoint [53] available online at https://sparql.omabrowser.org/sparql.
The class hierarchy of the OMA ontology. Ellipses indicate class labels, while arrows indicate the “rdfs:subClassOf ” property. Further details are available in [13]
Until here we have explored a few relatively simple examples in order to get familiar with the basics of Semantic Web technologies (URIs, RDF triples, and SPARQL). However, we can now introduce a more complex query that will better illustrate the expressivity of the SPARQL query language for accessing RDF stores—that is, for integrating and joining data across different databases.
Since all RDF stores structure data using the same standard model for data interchange, the main requirements in order to efficiently join multiple sources are:
-
1.
That they each expose data through a SPARQL endpoint that supports federation (SPARQL 1.1)
-
2.
That the sources share URIs or ontologies
This is the reason why already today we can jointly query, for example, OMA and UniProt—essentially, integrating the two databases by means of executing a federated SPARQL query.
To illustrate this, let us consider the following example: what are the human genes available in the OMA database that have a known association with leukemia? OMA does not contain any information related to diseases, however, UniProt does. In this case, since OMA already cross-references UniProt with the oma:xrefUniprot property, we can write the following federated SPARQL query, which will be running at the OMA SPARQL endpoint:
select distinct ?proteinOMA ?proteinUniProt where { service <http://sparql.uniprot.org/sparql> { ?proteinUniProt a up:Protein . ?proteinUniProt up:organism taxon:9606 . # Homo Sapiens ?proteinUniProt up:annotation ?annotation . # annotations of this protein entry ?annotation rdfs:comment ?text filter( regex(str(?text), "leukemia") ) # only those containing the text "leukemia" } ?proteinOMA a orth:Protein. ?proteinOMA oma:xrefUniprot ?proteinUniProt. }
We skip the details regarding the prefixes used in the example and focus on the new elements in the query. The main part to point out is the “service < http://sparql.uniprot.org/sparql >” block, delimited between the inner brackets. This enables using the SPARQL endpoint of UniProt remotely, as a service. Through this mechanism, the query will first fetch from UniProt all instances of proteins that are annotated with a text that contains “leukemia” (this is achieved by the filter keyword in the service block). Then, using the cross-reference oma:xrefUniprot property, the query will return all the equivalent entries from OMA. From here, the user can explore, either in the OMA browser or by further refining the SPARQL query, other properties of these proteins: for example, their orthologs in a given species available in the database. In Exercise D at the end of this chapter, we encourage the reader to try this out in the OMA SPARQL endpoint. Note that the same results can be obtained by writing this query in the UniProt SPARQL endpoint and referencing the OMA one as a service. For an overview of federation techniques for RDF data, we refer the reader to the survey [54].
The mechanisms illustrated so far, while indeed powerful for federating distinct databases, have a major drawback: they require the user to know the schema of the databases (otherwise, how would we know which properties to query in the previous examples?), and, more importantly, they require all users to be familiar with a technical query language, such as SPARQL. While very expressive, formulating such queries can quickly become overwhelming for non-programmer users. In the following, we will look at techniques that aim to overcome these limitations.
5 Ontology-Based Integration of Heterogeneous Data Stores
So far we have seen some of the alternatives available for storing biological data—relational databases and triple stores. In this section, we look at how these heterogeneous sources can be integrated and accessed in a unified, user-friendly manner that does not require knowledge of the location or structure of the underlying data nor of the technical language (SQL or SPARQL) used to retrieve the data. The architecture we present is inspired by work presented in [21], which focused strictly on keyword search in relational databases.
5.1 A System’s Perspective
We start with a bottom-up description of the layers that make up an integrated data access system, followed by a concrete example using the two bioinformatics databases introduced above: the orthology database OMA and the gene expression database Bgee.
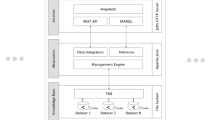
The main four layers of an integrated data access system, as shown in Fig. 9, are:
5.1.1 Base Data Layer
This represents the physical storage layer, where all the actual data, for example, experimental results, annotations, etc., are kept. Figure 9 illustrates only a few of the possible storage types, namely, relational databases, hierarchical data stores (e.g., HDF5), and RDF stores. At this low-level layer, the data are usually structured so as to optimize machine parameters, such as storage space, complexity of joins required to answer physical queries, etc. Therefore, it is not designed for human readability. Furthermore, tables, column names, or even IDs may not match any real terms. For example, the Bgee relational database uses the table name “anatEntity” to refer to the term “anatomic entity,” while others may be even further away from the original terms.
5.1.2 Data Model Layer
This layer is used to describe, at a higher level of abstraction, the data contained in the physical storage. Here, for example, original names for terms are recovered while also creating a mapping between these higher-level terms (“Anatomical Entity”) and their corresponding physical layer location (table “anatEntity” in schema Bgee). The data model layer can be viewed as the first semantic layer in the system, as it allows representing the actual terms referred to in the underlying physical storage while abstracting away the details of the actual structure of the physical storage. The data model layer can be understood as an ontology, however, only applicable to the level of an individual database.
5.1.3 Integration Layer
The integration layer performs a similar task to the data model layer, in that it defines a mapping between high-level concepts (“Anatomical Entity”) and all the occurrences where these concepts can be found in the physical storage (table “anatEntity” in schema Bgee, class “Anatomic Entity” in UniProt, etc.). In doing so, the integration layer also aligns the different data models, by defining which identifiers from one data model correspond to which ones from the others. In the case of biological databases, this is usually done by taking into account cross-references, which already exist between most databases, as we have seen in the SPARQL query in Sect. 5.
While the data model layer can be seen as a local ontology, the integration layer will serve as a global ontology. The integration layer can be queried using, for example, SPARQL. However, in order to get the results from the underlying sources, the SPARQL query needs be translated in the native query languages of the underlying sources (e.g., SQL for relational databases). This is achieved by using the mappings defined in the global ontology. For example, the keyword “expressed in” does not have a direct correspondence in Bgee, but it can be translated into an SQL procedure (in technical terms, it represents an SQL view of the data). Without going into details, at a high level, the property “gene A expressed in anatomic entity B” will be computed by looking at the number of experiments stored in the database, showing the expression of A in B. It is conceivable that in another database, which could also form part of the integrated system, this information is available explicitly. In this case the mapping would simply be a 1-to-1 correspondence to the property value stored in the database. The role of the integration layer is to capture all the occurrences where a certain concept (entity or property) can be found, along with a mapping for each of the occurrences, defining how information about this concept can be computed from the base data.
To summarize, the integration layer abstracts away the location and structure of data in the underlying sources, providing users a unified access through a global ontology. One of the drawbacks of this approach is that, in the lack of a presentation layer, such as a user-friendly query interface (e.g., a visual query builder or a keyword-based search interface), the data represented in the global ontology is accessible mainly through a technical query language, such as SPARQL. Therefore, in order to be able to access the data, users are required to become fluent in the respective query language.
It is worth at this point mentioning that most data integration systems available at the time of this writing only offer the three layers presented so far. Examples of such systems, generically denoted as ontology-based data access (OBDA) systems, are Ontop [55], Ultrawrap [56], or D2RQ [57].
5.1.4 Presentation Layer
The three layers presented so far already achieve data integration, but with a significant drawback, which is that the user is required to know a technical query language, such as SPARQL. The role of the presentation layer is to expose data from all integrated resources in an easy to access, user-friendly manner. The presentation layer abstracts away the structure of the integration layer and exposes data through a search interface that users (including non-programmers) are familiar with, such as keyword search [21, 58] or even full natural language search [59, 60].
The challenges in building the presentation layer are manyfold: first, human language is inherently ambiguous. As an example, let us assume a user asks: “Is the HBB gene expressed in the blood?” What does the user mean? The hemoglobin gene (HBB) in general? Or just in the human? The system should be proactive in helping the user clarify the semantics or intents of the question, before trying to compute the underlying SPARQL query. Second, the presentation layer should provide not only raw results but also an explanation—for example, what sources were queried, how many items from each source have been processed in order to generate the response, etc. This enables the user to validate the generated results or to otherwise continue refining the question. Third, the presentation layer must also rank the results according to some relevance metric, similarly to how search results are scored in Web search engines. Given that the number of results retrieved from the underlying sources can easily become overwhelming (e.g., searching for “HBB” in Bgee returns over 200 results), it is important that the most relevant ones are shown first.
From a technical point of view, the presentation layer maintains an index (i.e., the vocabulary) of all keywords stored in the lower layers, both data and metadata (descriptions, labels, etc.), such that each keyword in a user query can be mapped to existing data in the lower layers. An important observation is that the presentation layer highly relies on the quality of the annotations available in the lower layers. In the lack of human-readable labels and descriptions in the global ontology, the vocabulary collected by the presentation layer will miss useful terms that the user might search for. One way to detect and fix this problem is to always log user queries and improve the quality of the annotations “on demand,” whenever the queries cannot be solved due to missing items in the vocabulary. For a more extended discussion on the topic of labels and their role in the Semantic Web, refer to [61].
Finally, it is worth noting that none of these layers need to be centralized—indeed, even in the case of the integration layer, although its role is to build a common view of all data in the physical storage, it can be distributed across multiple machines, just as long as the presentation layer knows which machine holds which part of the unified view.
5.2 A Concrete Example: A Global Ontology to Unify OMA and Bgee
So far we have seen an abstract view of a system for data integration across heterogeneous databases. It is time to look at how this translates into a real-world example, using the Bgee relational database and the OMA RDF database.
The top part of Fig. 10, the terminological box, illustrates part of the global ontology (layer 3, integration layer) for the two databases, with most of the terms being part of OMA, except for Anatomic Entity, which is specific to Bgee. As mentioned previously, OMA extends the ORTH ontology, which is why the corresponding terms in the ontology are prefixed with “orth:.” The Gene concept can actually be found in both Bgee and OMA; therefore the global ontology will define mappings to both sources. As we can see in the ontology, the Gene is the common point that joins together OMA and Bgee. The gene IDs used in both databases are Ensembl IDs [62], stored in the ensemblGeneId string property. For example, the human hemoglobin gene, “HBB,” which we previously showed as an example entry in OMA, corresponds to the ENSG00000244734 Ensemble ID and can also be found in Bgee.
The lower part of Fig. 10, the assertional box, illustrates an example assertion—in this case, that the protein HUMAN22168 in OMA is orthologous to the protein HORSE13872 and that, furthermore, this protein is encoded by the gene with the Ensemble ID ENSG000001639936. Moreover, this gene is expressed in the brain (the Uberon ID for this being “UBERON:0000955”). The human-readable description is stored in the String literal label—as, for example, the name of the anatomic entity, “brain,” shown in the bottom-right corner in the figure. Without labels, much of the available data would not be easily searchable by a human user nor by an information retrieval system.
Note that with this sample ontology, we can already answer questions related to orthology and gene expression jointly, such as the first part of our introductory query: “What are the human-rat orthologs, expressed in the liver…?”. This question essentially refers to pairs of orthologous Genes (those in human and rat) and their expression in a given Anatomic Entity (the liver). Apart from the Species class, which is not explicitly shown, all of the information is already captured by the ontology in Fig. 10. A similar mechanism can be used to further extend this to UniProt (for instance, based again on gene IDs as the “join point,” or by using existing cross-references, as we have shown in the previous section), therefore enabling users to ask even more complex queries.
5.3 How to Link a Database with an Ontology?
One of the main challenges in implementing technologies for the Semantic Web was recognized from early on (see the study published in 2001 by Calvanese et al. [63]) to be the problem of integrating heterogeneous sources. In particular, one of the observations made was that integrating legacy data will not be feasible through a simple 1-to-1 mapping of the underlying sources into an integrative ontology (e.g., mapping all attributes of tables in relational databases to properties of classes in an ontology), but rather through more complex transformations, that map views of the data into elements of the global ontology [63].
To illustrate this with a concrete example, let us consider again the unified ontology for OMA and Bgee that we introduced in the previous section. Although Figure 10 shows properties such as “gene isExpressedIn” or “gene hasOrtholog,” this data is actually not explicitly stored in the underlying databases but rather needs to be computed on-the-fly based on the available data. For example, the “isExpressedIn” property can be computed based on the number of experiments which show the expression of a gene in a certain anatomic entity in Bgee. Deciding the exact threshold for when a gene is considered as “expressed” according to the data available is not straightforward and needs to be agreed upon by domain specialists. Therefore, the integration layer will also serve to enrich the data available in the underlying layers, by defining new concepts based on this data (e.g., the presence or absence of gene expression in an anatomic entity).
At this point it is worth clarifying an important question: why are mappings necessary? Why is it not enough to replicate the data in the different underlying formats into a single, uniform way (e.g., translate all RDB data into RDF)? The answer is that not only would such a translation require a lot of engineering effort, but more importantly, it would transform the data from a format that is highly optimized for data access, into a format that is optimized for different purposes (data integration and reasoning). Querying relational databases still is, today, the most efficient means of accessing very large quantities of structured data. Transforming all of it into RDF would in many cases mean downgrading the overall performance of the system. In some cases storing RDF data in the relational format was proven to be more efficient [64].
So how are mappings then created? One of the main mechanisms to achieve this is currently the W3C standard R2RML, available as a W3C recommendation online [65]. R2RML enables mapping relational data to the RDF model, as chosen by the programmer. For a concrete example of how mappings can be defined and what are the advantages of this approach, we refer the reader to [66]. A mapping essentially defines a view of the data, which is a query (in this case, an SQL query) that allows retrieving a relevant portion of the underlying data, in order to answer a higher-level question (e.g., what is “expressed in”?). The materialization of this query (the answer) will be returned in RDF format, on demand, according to the mapping. This avoids duplicating or translating data in advance from the underlying relational database into RDF until it is really needed, in order to answer a user query.
For a discussion regarding the limitations of R2RML and alternative approaches to define mappings from relational data to RDF, we refer the reader to the survey [67].
5.4 Putting Things Together
So far we have seen how individual sources can be represented into a single, unified ontology, and we had a high-level view of a data access system that enables users to ask queries and get responses in a unified way, without knowledge of where data is located or how it is structured. In this section we finally look at how all of these components can work together in answering natural language queries on biological databases. Although there are multiple alternatives to natural language interfaces, including visual query interfaces or keyword-based search interfaces, it has been shown that natural language interfaces are the most appropriate means to query Semantic Web data for non-technical end-users [68]. As a consequence, natural language querying, based on Semantic Web technologies, is currently one of the active areas of research, examples of recent systems implementing an ontology-based natural language interface including the Athena [59] and TRDiscover [60] systems.
First, recall the user question we formulated in the beginning of this chapter: “What are the human-rat orthologs, expressed in the liver, that are associated with leukemia?” Let us assume the resources at hand to answer this question are the biological databases OMA, Bgee, and UniProt. The four main steps required to translate the natural language question into the underlying query languages of OMA, Bgee, and UniProt will be:
-
(a)
Identify entities in the query
This is the natural language processing step that extracts the main concepts the user is interested in, based on the keywords of the input query: orthologs, human, rat, expressed, liver, associated, and leukemia.
-
(b)
Identify matches of the entities in the integrative ontology
The extracted keywords will be searched for in the vocabulary of the presentation layer, resulting in one or multiple URIs, given that a keyword can match multiple concepts. For example, the keyword “orthologs” can match either the entity “OrthologCluster” or the property “hasOrtholog” of a gene in OMA. The index of the presentation layer will also return the location the URI originates from (OMA or Bgee or UniProt).
-
(c)
Construct subqueries for each of the matches
The extracted URIs will be used to construct subqueries on each of the underlying data sources. This step requires translating the original query into the native language of each underlying database, with specific mechanisms for each type of database (relational or triple store). At a high level, the translation process involves finding the minimal sub-schema (or subgraph in the case of RDF data) that covers all the keywords matched from the input query. Taking the example previously shown in Fig. 10, the minimal subgraph that contains “orthologs” and “expressed” will essentially contain only two nodes of the entire graph: Gene (which is both the domain and the range of the “hasOrtholog” property in the Orthology Ontology) and AnatomicEntity (which is the range of the “isExpressedIn” property in the Bgee ontology). All the unknowns of the query (e.g., which ortholog genes) are replaced by variables. The final subqueries for OMA and Bgee might therefore (informally) look like this:
OMA: select ?gene1 ?gene2 where { ?protein1 a Protein. ?protein1 inTaxon “Homo sapiens”. ?protein1 isEncodedBy ?gene1. ?protein1 hasOrtholog ?protein2. ?protein2 inTaxon “Rattus norvegicus”. ?protein2 isEncodedBy ?gene2. }
Note that we have simplified the actual query for readability purposes (using the literals “Homo sapiens” and “Rattus norvegicus” instead of their corresponding URIs). This subquery will cover the keywords: ortholog, human, and rat. Notice that the query should return genes, not proteins, because the join point between OMA and Bgee is the Gene class.
Bgee: select ?gene where { ?gene a Gene. ?gene isExpressedIn ?anatomicEntity. ?anatomicEntity rdfs:label “liver”. }
This subquery will therefore cover the expressed and liver keywords. The final step will be then to get the similar subquery for UniProt (which we omit here for brevity) and to compute the joint result, namely, the intersection between all the sets returned by the subqueries.
-
(d)
Join the results from each of the subqueries
This final step is essential in keeping the performance of the system to an acceptable level. Joining (federating) the results of several subqueries into a unified result is not an easy task and requires a careful ordering of the operations from all subqueries. To understand this problem, let us consider again our example and try to see how many results each of the subqueries will return. First, if we take a look at the OMA browser and try to find all orthologs between human and rat, this will amount to more than 21,000 results. However, is the user really interested in all of them? Certainly not, as the input query shows—the user is only interested in a small fraction of the orthologs, namely, those that are expressed in the liver and have an association with leukemia (according to the data stored in Bgee and UniProt). How many are these? If we now refer to UniProt and look for the disease leukemia, we will find that there are only 20 entries which illustrate the association with this disease. Clearly, getting only the orthologs of these 20 entries will be much more efficient than retrieving all 21,000 pairs from OMA first and then removing most of them to only keep relevant ones.
However, note that in this case, we only know this information because we constructed the queries and tried them out by hand first. How should the system estimate the number of results (i.e., the cardinality of each subquery) in advance? This question has been an active area of research for a long time. Some of the methods used to tackle this problem are either to precompute statistics regarding the number of results available in different tables of the underlying sources [69] or to use statistics regarding previously asked queries to optimize the new ones, for example, via statistical machine learning [70]. In the first case, we would, for instance, store the individual counts of different orthologous pairs while also keeping statistics about diseases if we expect these types of questions to be asked frequently, whereas in the second case, we would simply look at the number of results similar subqueries generated in the past, to optimize which results to fetch first. For a recent study of optimization methods for federated SPARQL queries, see [71].
-
(e)
Present the user the final results
Finally, the joined results are returned to the user, along with an explanation regarding the constructed query and the entities that were matched in order to construct it. In this way, the user has the opportunity to validate the correctness of the answer or otherwise to further refine the question.
For a more in-depth discussion regarding natural language query interfaces in ontology-based data access systems, we refer the reader to Athena [59] and TRDiscover [60].
6 Timeline of Semantic Web Technologies and Ontology-Based Data Integration in Life Sciences
The field of life sciences has been an early adopter of Semantic Web technologies, due to the need of interoperability and integration of biological data spread across different databases. In this section, we provide a brief timeline (see Fig. 11), including the example ontologies introduced in this chapter.
-
1995: Davidson et al. [72] suggest basic steps to integrate bioinformatics data (common data model, match semantically related objects, schema integration, transform data into federated database, match semantically equivalent data).
-
2000: TAMBIS (Transparent Access to Multiple Bioinformatics Information Sources) [73] proposes a unified ontology covering many aspects of the bioinformatics knowledge space.
-
2000: The “Gene Ontology—a tool for the unification of biology” [37] is the first significant milestone in unifying diverse biological databases, focusing on gene functions. Even before the publication of the Semantic Web paper by Tim Berners Lee (in the following year), the GO highlighted the benefits of controlled vocabularies and standardized naming, both precursors of Semantic Web technologies, which were adopted in the GO in the year 2002 [74]. Today it is, arguably, the most comprehensive resource of computable knowledge regarding gene functions and products.
-
2001: Launch of the BioMoby project [75] providing a unified registry of Web services for life scientists using a consensus-driven approach. It listed, for instance, all services converting gene names to GO terms or all databases accepting GO terms. The registry is currently no longer maintained.
-
2003: A Nature Reviews Genetics article on Integrating Biological Databases [76] highlights the “database-surfing” problem (i.e., the time-consuming process of manually visiting multiple databases to answer complex biological research questions) and argues for standardized naming of biological objects to overcome the problem. Link integration, view integration, and data warehousing are proposed for data integration. Arguably, link integration has since become the most adopted solution.
-
2003: Launch of UniProt [77] by the UniProt Consortium, a collaboration between the Swiss Institute of Bioinformatics (SIB), the European Bioinformatics Institute (EBI), and the Protein Information Resource (PIR). UniProt is the world’s most comprehensive freely accessible resource on protein sequences and functional annotation. Since 2008 the data is published in RDF, and since 2013 a SPARQL endpoint is provided [78].
-
2004: The first International Workshop on Data Integration in the Life Sciences, held in Leipzig, promotes “a Bioinformatics Semantic Web” and highlights solutions for heterogeneous data integration. The workshop continues to be held every year, and its proceedings (e.g., [79]) provide a good overview of advances in the field.
-
2005: The W3C Consortium launches the Semantic Web Health Care and Life Sciences Interest Group (HCLS IG) to develop the use of Semantic Web technologies to improve health care and life sciences research. Today, the HCLS Linked Data Guide [80] provides best practices for publication of biological Linked Data on the Web.
-
2006: The OBO Foundry [40] establishes principles for ontology development and evolution to support biomedical data integration through a suite of orthogonal interoperable reference ontologies.
-
2006: Publication of the Ontology Lookup Service (OLS), a repository for biomedical ontologies with the aim to provide a single point of access (with controlled vocabulary queries) to the latest ontology versions. It allows interactive browsing, as well as programmatic access [81].
-
2007: Launch of the National Center for Biomedical Ontology (NCBO) BioPortal [82], a web portal to biomedical ontologies. OBO ontologies are a central component. The portal started with 50 ontologies; to date it is the most comprehensive repository with currently 852 biomedical ontologies and more than eight million classes.
-
2008: Launch of the BioMoby Consortium [83] and the first release of the BioMoby Semantic Web Service, at the time providing interoperable access to over 1400 bioinformatics resources worldwide.
-
2008: BioGateway [84] provides a single SPARQL entry point to all OBO candidate ontologies, the GO annotation files, the SWISS-PROT protein set, the NCBI taxonomy, and several in-house ontologies.
-
2008: The Briefings in Bioinformatics journal launches a special issue dedicated to Database Integration in Life Sciences [85], acknowledging the major challenge of integrating data scattered over millions of publications and thousands of heterogeneous databases.
-
2008: Bio2RDF [86] applies Semantic Web technology to various publicly available databases (converting them into RDF format and linking with normalized URIs and a common ontology). Updates continue to be provided for increased interoperability among bioinformatics databases [87, 88].
-
2009: Briefings in Bioinformatics publishes a review on Biological Knowledge Management [89], highlighting the transforming role of ontologies and Semantic Web technologies in enabling knowledge representation and extraction from heterogeneous bioinformatics databases.
-
2010: NCBO launches a SPARQL endpoint, available at http://sparql.bioontology.org/.
-
2012: Publication of a survey highlighting the benefits of integration using Semantic Web technologies in the field of Integrative Biology [90].
-
2016: Publication of the Orthology Ontology [13].
7 Conclusions and Outlook
Data integration is arguably one of the most important enablers of new scientific discoveries, given that research data is currently growing at an unprecedented rate. This is especially true in the case of biological databases. While data integration poses many challenges, the emergence of standards, integrative ontologies, as well as the availability of cross-references between many of the biological databases make the problem easier to tackle. This chapter has provided a brief introduction to the methods that can be used to integrate heterogeneous databases using Semantic Web technologies while also providing a concrete example of achieving this goal for three well-known existing biological databases: OMA, Bgee, and UniProt.
Although there would be many more aspects to cover and much of the work for achieving wide-scale data integration still remains to be done, we would like to end this chapter by reinforcing the following conclusion, extracted from a study of Biological Ontologies for Biodiversity Knowledge Discovery [91]:
We hope that current work will spur interest and feedback from scientists and bioinformaticians who see data integration, interoperability, and reuse as the solution to bringing the past 300 years of biological exploration of the planet into currency for science and society.
8 Exercises
-
A. Querying UniProt with SPARQL
The goal of this warm-up exercise is to get familiar with a SPARQL endpoint and to write your first SPARQL query. For this purpose, open the link to the UniProt SPARQL endpoint, http://sparql.uniprot.org/ in a Web browser. How many entries do you think are available in UniProt? To find out, simply check the bottom-left corner of the Web page—you will notice that the total number of triples is always kept up to date there. How many of these entries describe proteins? To find out, try running the following SPARQL query that counts all instances of the database that belong to the protein class. What is the result?
PREFIX up:<http://purl.uniprot.org/core/> SELECT (count(?protein) as ?count) WHERE { ?protein a up:Protein. }
Notice that the UniProt SPARQL web page includes many examples on the right-hand side—in order to get more familiar with UniProt and SPARQL, try further some of the sample queries provided there.
-
B. Exploring Biological Ontologies Through Keyword Search in the Ontology Lookup Service
We have seen in Sect. 3.6 an example assertion about the “HBB” gene in the human, including the following triple:
oma:PROTEIN_HUMAN04027 obo:RO_0002162 <http://www.uniprot.org/taxonomy/9606 > .
This triple essentially asserts that the gene is located in the Homo sapiens taxon. However, as a regular user, how could you know what the URIs for “in taxon” and Homo sapiens are? One of the possible ways to get these identifiers is by searching for the keywords of interest in the Ontology Lookup Service (OLS). To do this, go to the Web page of the service https://www.ebi.ac.uk/ols/index , and try to enter first “in taxon”. What is the result? Try also Homo sapiens. What about “human”?
-
C. Querying OMA with SPARQL
Recall from Sect. 3.6 the sample query we presented for retrieving the description of the human hemoglobin gene from OMA. We provide it in a more explicit form here:
SELECT ?description WHERE { ?protein oma:geneName "HBB". ?protein < http://bioontology.org/ontologies/biositemap.owl#description > ?description. }
First try to think about possible information that is missing from this query. For example, is this query guaranteed to return a single result (remember we are using an orthology database)?
Try to look again at how the human “HBB” protein is defined in Sect. 3. Then, try to run the SPARQL query as-is in the OMA SPARQL endpoint: https://sparql.omabrowser.org/sparql . What do you get? What is the reason? Try to print out more information about the protein, not just its description. For example, add another triple pattern to capture the oma:hasOMAId property value as well (don’t forget to add it to the selected variables in the first line!), perhaps also the taxon ID in UniProt. What can you deduce? Can you correct the query so that it only gets the description we were originally interested in?
-
D. Federated Queries Using SPARQL (OMA and UniProt)
In Sect. 4 we presented an example Federated Query using the SPARQL endpoint of OMA and the remote SPARQL endpoint of UniProt, as a service. We recall the query here:
prefix up:<http://purl.uniprot.org/core/> prefix taxon:<http://purl.uniprot.org/taxonomy/> select distinct ?proteinOMA ?proteinUniProt where { service <http://sparql.uniprot.org/sparql> { ?proteinUniProt a up:Protein . ?proteinUniProt up:organism taxon:9606 . # Homo Sapiens ?proteinUniProt up:annotation ?annotation . # annotations of this protein entry ?annotation rdfs:comment ?text filter( regex(str(?text), "leukemia") ) # only those containing the text "leukemia" } ?proteinOMA a orth:Protein. ?proteinOMA oma:xrefUniprot ?proteinUniProt. }
Try running this query in the OMA SPARQL endpoint, https://sparql.omabrowser.org/sparql . You might need to wait a couple of minutes to get the remote results. Next, try to look at the examples provided in the right side of the page to see how to get more properties of the proteinOMA variable—for example, try getting the description or the OMA ID. Next, try modifying this query so that it can run in the UniProt SPARQL endpoint, invoking the OMA one as a service. Remember to get the relevant prefixes and define them in the header of the query first (“oma,” “orth”). You can get these by looking at “Namespace prefixes” in the OMA SPARQL Web page. Finally, test your modifications using UniProt, http://sparql.uniprot.org/ .
Abbreviations
- ABox:
-
Assertional box
- Bgee:
-
dataBase for Gene Expression Evolution, https://bgee.org/
- FK:
-
Foreign key in a relational database
- HBB:
-
Hemoglobin unit beta gene
- IRI:
-
Internationalized Resource Identifier
- OBDA:
-
Ontology-based data access
- OMA:
-
Orthologous Matrix, a database for the inference of orthologs among complete genomes.—https://omabrowser.org, SPARQL endpoint: https://sparql.omabrowser.org/sparql
- PK:
-
Primary key in a relational database
- PK-FK:
-
Primary key-foreign key relationship; enables joining two tables in a relational database
- RDB:
-
Relational database
- RDF:
-
Resource Description Framework
- SODA:
-
Search Over Relational Databases [21]
- SQL:
-
Structured Query Language
- SPARQL:
-
SPARQL Protocol and RDF Query Language
- TBox:
-
Terminological box
- URI:
-
Uniform Resource Identifier
References
Mole B (2004) The gene sequencing future is here. http://www.sciencenews.org/article/gene-sequencing-future-here. Accessed 15 Feb 2018
Stephens ZD, Lee SY, Faghri F et al (2015) Big data: astronomical or genomical? PLoS Biol 13(7):e1002195. https://doi.org/10.1371/journal.pbio.1002195
Altenhoff AM, Škunca N, Glover N et al (2014) The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res 43(D1):D240–D249. https://doi.org/10.1093/nar/gku1158
Waterhouse RM, Tegenfeldt F, Li J et al (2012) OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res 41(D1):D358–D365. https://doi.org/10.1093/nar/gks1116
Powell S, Szklarczyk D, Trachana K et al (2011) eggNOG v3. 0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40(D1):D284–D289
Bastian F, Parmentier G, Roux J et al (2008) Bgee: integrating and comparing heterogeneous transcriptome data among species. In: International Workshop on Data Integration in the Life Sciences. Springer, Berlin, pp 124–131
UniProt Consortium (2014) UniProt: a hub for protein information. Nucleic Acids Res 43(D1):D204–D212. https://doi.org/10.1093/nar/gku989
Codd EF (1970) A relational model of data for large shared data banks. Commun ACM 13(6):377–387. https://doi.org/10.1145/362384.362685
W3Schools. Online SQL Tutorial. https://www.w3schools.com/sql/sql_intro.asp. Accessed 15 Feb 2018
Beaulieu A (2009) Learning SQL: master SQL fundamentals. O’Reilly Media, Inc, Sebastopol, CA
Fehily C (2014) SQL (database programming). Questing Vole Press, Pacific Grove, CA. (2015 Edition)
Teodoro D, Pasche E, Wipfli R et al (2009) Integration of biomedical data using federated databases. Swiss Medical Informatics, Muttenz
Fernández-Breis JT, Chiba H, del Carmen L-GM et al (2016) The orthology ontology: development and applications. J Biomed Semant 7(1):34. https://doi.org/10.1186/s13326-016-0077-x
Self-join incurs more I/O activities and increases locking overhead (2013) http://sqltouch.blogspot.ch/2013/07/self-join-incurs-more-io-activities-and.html. Accessed 15 Feb 2018
Sadalage PJ, Fowler M (2012) NoSQL distilled: a brief guide to the emerging world of polyglot persistence. Pearson Education, Upper Saddle River, NJ
Hunger M, Boyd R, Lyon W (2016) RDBMS & Graphs: why relational databases aren’t always enough. https://neo4j.com/blog/rdbms-graphs-why-relational-databases-arent-enough/. Accessed 15 Feb 2018
Stockinger K, Bödi R, Heitz J et al (2017) ZNS-Efficient query processing with ZurichNoSQL. Data Know Eng 112:38–54
Erling O, Mikhailov I (2009) RDF support in the virtuoso DBMS. In: Networked knowledge-networked media. Springer, Berlin, pp 7–24. https://doi.org/10.1007/978-3-642-02184-8_2
Berners-Lee T, Hendler J, Lassila O (2001) The semantic web. Sci Am 284(5):34–43
Kimball R, Ross M, Mundy J et al (2015) The kimball group reader: relentlessly practical tools for data warehousing and business intelligence remastered collection. John Wiley & Sons, New York, NY
Blunschi L, Jossen C, Kossmann D et al (2012) Soda: generating sql for business users. Proc VLDB Endow 5(10):932–943
Bio-SODA: enabling complex, semantic queries to bioinformatics databases through intuitive searching over data (2017) https://www.zhaw.ch/no_cache/en/research/people-publications-projects/detail-view-project/projekt/3066/. Accessed 15 Feb 2018
Lane L, Argoud-Puy G, Britan A et al (2011) neXtProt: a knowledge platform for human proteins. Nucleic Acids Res 40(D1):D76–D83. https://doi.org/10.1093/nar/gkr1179. Sparql endpoint. Available at: https://sparql.nextprot.org/
Li W, Cowley A, Uludag M et al (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43(W1):W580–W584. https://doi.org/10.1093/nar/gkv279. RDF data. Available at: https://www.ebi.ac.uk/rdf
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide protein data bank. Nat Struct Mol Biol 10(12):980. RDF data. Available at https://pdbj.org/help/rdf
Wilkinson MD, Dumontier M, Aalbersberg IJ et al (2016) The FAIR guiding principles for scientific data management and stewardship. Sci Data 3:160018. https://doi.org/10.1038/sdata.2016.18
Bizer C, Heath T, Berners-Lee T (2009) Linked data-the story so far. Int J Semant Web Inf Syst 5(3):1–22
Domingue J, Fensel D, Hendler JA (eds) (2011) Handbook of semantic web technologies. Springer Science & Business Media, New York, NY
Hitzler P, Krotzsch M, Rudolph S (2009) Foundations of semantic web technologies. CRC press, Boca Raton, FL
Patel-Schneider PF (2005) A revised architecture for semantic web reasoning. In: International Workshop on Principles and Practice of Semantic Web Reasoning. Springer, Berlin, pp 32–36
Bratt S (2007) Semantic Web, and Other Technologies to Watch. https://www.w3.org/2007/Talks/0130-sb-W3CTechSemWeb/#(24) . Accessed 15 Feb 2018
URN syntax, RFC2141 (1997) https://tools.ietf.org/html/rfc2141. Accessed 15 Feb 2018
URI Syntax, RFC3986 (2005) https://tools.ietf.org/html/rfc3986. Accessed 15 Feb 2018
RDF 1.1 Primer (2014) https://www.w3.org/TR/rdf11-primer/. Accessed 15 Feb 2018
Studer R, Benjamins VR, Fensel D (1998) Knowledge engineering: principles and methods. Data Know Eng 25(1-2):161–197
Baader F (ed) (2003) The description logic handbook: theory, implementation and applications. Cambridge university press, Cambridge
Ashburner M, Ball CA, Blake JA (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25. https://doi.org/10.1038/75556
Gene Ontology Consortium (2014) Gene ontology consortium: going forward. Nucleic Acids Res 43(D1):D1049–D1056. https://doi.org/10.1093/nar/gku1179
Dessimoz C, Gabaldón T, Roos DS et al (2012) Toward community standards in the quest for orthologs. Bioinformatics 28(6):900–904. https://doi.org/10.1093/bioinformatics/bts050
Smith B, Ashburner M, Rosse C et al (2007) The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25(11):1251
Federhen S (2011) The NCBI taxonomy database. Nucleic Acids Res 40(D1):D136–D143. https://doi.org/10.1093/nar/gkr1178
McBride B (2004) The resource description framework (RDF) and its vocabulary description language RDFS. In: Handbook on ontologies. Springer, Berlin, pp 51–65
OWL Web Ontology Language Overview (2004) https://www.w3.org/TR/owl-features/. Accessed 15 Feb 2018
Motik B, Patel-Schneider PF, Parsia B, Bock C, Fokoue A, Haase P, Hoekstra R, Horrocks I, Ruttenberg A, Sattler U, Smith M (2009) OWL 2 web ontology language: structural specification and functional-style syntax. W3C Recomm 27(65):159
OWL Web Ontology Language semantics and abstract syntax (2004) W3C Recommendation, 10. https://www.w3.org/TR/owl-semantics/. Accessed 15 Feb 2018
W3C Owl Working Group (2012) OWL 2 web ontology language document overview. https://www.w3.org/TR/owl2-overview/. Accessed 15 Feb 2018
Dean M, Schreiber G, Bechhoffer S, et al (2004) OWL web ontology language reference. W3C Recommendation. https://www.w3.org/TR/owl-ref/. Accessed 15 Feb 2018
Prud’hommeaux E, Carothers G (2014) RDF 1.1 Turtle: terse RDF triple language. W3C recommendation. http://www.w3.org/TR/2014/REC-turtle-20140225/. The latest edition is available at http://www.w3.org/TR/turtle/
World Wide Web Consortium (2014) RDF 1.1 XML Syntax. The latest edition is available at https://www.w3.org/TR/rdf-syntax-grammar/
World Wide Web Consortium (2014) JSON-LD 1.0: a JSON-based serialization for linked data. The latest edition is available at https://www.w3.org/TR/json-ld/
SPARQL Query Language for RDF (2008) https://www.w3.org/TR/rdf-sparql-query/. Accessed 15 Feb 2018
Lohmann S, Link V, Marbach E et al (2014) WebVOWL: web-based visualization of ontologies. In: International Conference on Knowledge Engineering and Knowledge Management. Springer, Cham, pp 154–158
Altenhoff AM, Glover NM, Train CM et al (2017) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46(D1):D477–D485
Rakhmawati NA, Umbrich J, Karnstedt M et al (2013) A comparison of federation over SPARQL endpoints frameworks. In: International Conference on Knowledge Engineering and the Semantic Web. Springer, Berlin, pp 132–146
Calvanese D, Cogrel B, Komla-Ebri S et al (2017) Ontop: answering SPARQL queries over relational databases. Semant Web 8(3):471–487
Sequeda JF, Miranker DP (2013) Ultrawrap: SPARQL execution on relational data. Web Semant 22:19–39
Bizer C, Seaborne A (2004) D2RQ-treating non-RDF databases as virtual RDF graphs. In: Proceedings of the 3rd international semantic web conference (ISWC2004). Springer, New York, NY
Gasteiger E, Gattiker A, Hoogland C et al (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788. https://doi.org/10.1093/nar/gkg563
Saha D, Floratou A, Sankaranarayanan K et al (2016) Athena: an ontology-driven system for natural language querying over relational data stores. Proc VLDB Endow 9(12):1209–1220. https://doi.org/10.14778/2994509.2994536
Song D, Schilder F, Smiley C et al (2015) TR Discover: a natural language interface for querying and analyzing interlinked datasets. In: International Semantic Web Conference, 2. Springer, Cham
Ell B, Vrandečić D, Simperl E (2011) Labels in the web of data. Semant Web 2011:162–176. https://doi.org/10.1007/978-3-642-25073-6_11
Kinsella RJ, Kähäri A, Haider S et al (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011:bar030. https://doi.org/10.1093/database/bar030
Calvanese D, De Giacomo G, Lenzerini M (2001) Ontology of integration and integration of ontologies. Descr Logics 49(10-19):30
Bornea MA, Dolby J, Kementsietsidis A et al (2013) Building an efficient RDF store over a relational database. In: Proceedings of the 2013 ACM SIGMOD International Conference on Management of Data. ACM, Washington, DC, pp 121–132
R2RML: RDB to RDF mapping language (2012) https://www.w3.org/TR/r2rml/. Accessed 15 Feb 2018
Sequeda JF (2016) Integrating relational databases with the Semantic Web. IOS Press, Amsterdam
Michel F, Montagnat J, Zucker CF (2013) A survey of RDB to RDF translation approaches and tools. Dissertation. Inria Sophia Antipolis
Kaufmann E, Bernstein A (2007) How useful are natural language interfaces to the semantic web for casual end-users? In: The Semantic Web. Springer, Berlin, pp 281–294
Leis V, Gubichev A, Mirchev A et al (2015) How good are query optimizers, really? Proc VLDB Endow 9(3):204–215
Wu W, Chi Y, Zhu S et al (2013) Predicting query execution time: are optimizer cost models really unusable? In: Data Engineering (ICDE), 2013 IEEE 29th International Conference on 2013 Apr 8. IEEE, New York, NY, pp 1081–1092
Montoya G, Skaf-Molli H, Hose K (2017) The Odyssey approach for optimizing federated SPARQL queries. In: International Semantic Web Conference. Springer, Cham, pp 471–489
Davidson SB, Overton C, Buneman P (1995) Challenges in integrating biological data sources. J Comput Biol 2(4):557–572. https://doi.org/10.1089/cmb.1995.2.557
Stevens R, Baker P, Bechhofer S et al (2000) TAMBIS: transparent access to multiple bioinformatics information sources. Bioinformatics 16(2):184–186
Magkanaraki A, Alexaki S, Christophides V et al (2002) Benchmarking rdf schemas for the semantic web. In: International Semantic Web Conference. Springer, Berlin, pp 132–146
Wilkinson MD, Links M (2002) BioMOBY: an open source biological web services proposal. Brief Bioinform 3(4):331–341. https://doi.org/10.1093/bib/3.4.331
Stein LD (2003) Integrating biological databases. Nat Rev Genet 4(5):337. https://doi.org/10.1038/nrg1065
Apweiler R, Bairoch A, Wu CH (2004) Protein sequence databases. Curr Opin Chem Biol 8(1):76–80. https://doi.org/10.1016/j.cbpa.2003.12.004
UniProt Consortium (2012) Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res 41(D1):D43–D47. https://doi.org/10.1093/nar/gks1068
Da Silveira M, Pruski C, Schneider R (2017) Data integration in the life sciences. Springer International Publishing AG, Cham
HCLS Linked Data Guide (2012) https://www.w3.org/2001/sw/hcls/notes/hcls-rdf-guide/. Accessed 15 Feb 2018
Côté RG, Jones P, Apweiler R et al (2006) The ontology lookup service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics 7(1):97. https://doi.org/10.1186/1471-2105-7-97
Salvadores M, Alexander PR, Musen MA et al (2013) BioPortal as a dataset of linked biomedical ontologies and terminologies in RDF. Semant Web 4(3):277–284. https://doi.org/10.3233/SW-2012-0086
BioMoby Consortium (2008) Interoperability with Moby 1.0—it’s better than sharing your toothbrush! Brief Bioinform 9(3):220–231. https://doi.org/10.1093/bib/bbn003
Antezana E, Blondé W, Egaña M et al (2009) BioGateway: a semantic systems biology tool for the life sciences. BMC Bioinformatics 10(10):S11. https://doi.org/10.1186/1471-2105-10-S10-S11
Database Integration in the Life Sciences (2008) Briefings in Bioinformatics, Special Issue 9(6). https://academic.oup.com/bib/issue/9/6
Belleau F, Nolin MA, Tourigny N et al (2008) Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J Biomed Inform 41(5):706–716. https://doi.org/10.1016/j.jbi.2008.03.004
Callahan A, Cruz-Toledo J, Ansell P et al (2013) Bio2RDF release 2: improved coverage, interoperability and provenance of life science linked data. In: Extended Semantic Web Conference. Springer, Berlin, pp 200–212. https://doi.org/10.1007/978-3-642-38288-8_14
Dumontier M, Callahan A, Cruz-Toledo J et al (2014) Bio2RDF release 3: a larger connected network of linked data for the life sciences. In: Proceedings of the 2014 International Conference on Posters & Demonstrations Track
Antezana E, Kuiper M, Mironov V (2009) Biological knowledge management: the emerging role of the Semantic Web technologies. Brief Bioinform 10(4):392–407. https://doi.org/10.1093/bib/bbp024
Chen H, Yu T, Chen JY (2012) Semantic web meets integrative biology: a survey. Brief Bioinform 14(1):109–125. https://doi.org/10.1093/bib/bbs014
Walls RL, Deck J, Guralnick R et al (2014) Semantics in support of biodiversity knowledge discovery: an introduction to the biological collections ontology and related ontologies. PLoS One 9(3):e89606
Acknowledgments
We would like to thank all the members of the Bio-SODA SNF project at ZHAW Zurich University of Applied Sciences, at the University of Lausanne (UNIL) and at the SIB Swiss Institute of Bioinformatics for their valuable feedback: Adrian Altenhoff (UNIL, SIB), Maria Anisimova (ZHAW, SIB), Christophe Dessimoz (UNIL, SIB), Frederic Bastian (UNIL, SIB), Heinz Stockinger (SIB), Marc Robinson-Rechavi (UNIL, SIB), and Erich Zbinden (ZHAW, SIB). This work was supported by the Swiss National Science Foundation (SNSF) funded under grant NRP 75 Big Data.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this protocol
Cite this protocol
Sima, A.C., Stockinger, K., de Farias, T.M., Gil, M. (2019). Semantic Integration and Enrichment of Heterogeneous Biological Databases. In: Anisimova, M. (eds) Evolutionary Genomics. Methods in Molecular Biology, vol 1910. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9074-0_22
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9074-0_22
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9073-3
Online ISBN: 978-1-4939-9074-0
eBook Packages: Springer Protocols