Abstract
Expression and purification of turkey coronavirus (TCoV) nucleocapsid (N) protein from a prokaryotic expression system as histidine-tagged fusion protein are presented in this chapter. Expression of histidine-tagged fusion N protein with a molecular mass of 57 kDa is induced with isopropyl β-d-1-thiogalactopyranoside (IPTG). The expressed N protein inclusion body is extracted and purified by chromatography on nickel-agarose column to near homogeneity. The protein recovery can be 10 mg from 100 ml of bacterial culture. The purified N protein is a superior source of TCoV antigen for antibody-capture ELISA for detection of antibodies to TCoV.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Turkey coronavirus (TCoV) is the cause of an acute and highly contagious enteric disease affecting turkeys of all ages. The disease is severe in 1- to 4-week-old turkey poults [1]. Turkey flocks that recover from natural or experimental coronaviral enteritis may develop lifelong immunity [2]. TCoV has been recognized as an important pathogen of young turkeys. TCoV infection causes significant economic losses in the turkey industry due to poor feed conversion and uneven growth. Outbreaks of TCoV enteritis in turkey poults remain as a threat to the turkey industry.
In order to rapidly diagnose and effectively control turkey coronaviral enteritis , development of an antibody-capture enzyme-linked immunosorbent assay (ELISA ) for detecting antibodies to TCoV is essential. Development of ELISA for detection of TCoV infection requires large amounts of TCoV antigen . Molecular cloning and expression of TCoV N protein were carried out for preparation of large quantities of highly purified viral proteins.
Coronavirus is an enveloped and positive-stranded RNA virus that possesses three major structural proteins including a predominant phosphorylated nucleocapsid (N) protein, peplomeric glycoprotein, and spike (S) protein that makes up the large surface projections of the virion, and membrane protein (M) [3, 4]. The N protein is abundantly produced in coronavirus-infected cells and is highly immunogenic. The N protein binds to the viral genomic RNA and composes the structural feature of helical nucleocapsid.
The N protein is a preferred choice for developing a group-specific serologic assay because of highly conserved sequence and antigenicity. The nucleocapsid protein s of various RNA viruses, such as mumps, rabies, vesicular stomatitis, measles, Newcastle disease, and infectious bronchitis (IBV) viruses, have been used as the coating antigens in diagnostic ELISA [5–10]. Prokaryotic expression is an economic and convenient system to prepare large amount of pure recombinant protein. In addition, the antigenic integrity of N protein expressed in prokaryotic system is expected to be maintained due to the lack of glycosylation. This chapter describes expression and purification of TCoV N protein with a prokaryotic system for preparation of a large quantity of highly purified viral protein, which can be used as coating antigen for antibody-capture ELISA for serologic diagnosis of TCoV infection [11, 12].
2 Materials
2.1 Construction of TCoV N Gene in the Expression Vector pTriEx
-
1.
Virus source (see Note 1 ).
-
2.
RNApure reagent (GenHunter, Nashville, TN, USA).
-
3.
Chloroform.
-
4.
Isopropanol.
-
5.
70 % Ethanol.
-
6.
Diethyl-pyrocarbonate (DEPC)-treated sterile double-distilled water (DEPC-H2O).
-
7.
SuperScript III first-strand synthesis system for RT-PCR kit (Life Technologies/Invitrogen, Carlsbad, CA, USA).
-
8.
RNaseOUT, a recombinant RNase inhibitor (Life Technologies/Invitrogen).
-
9.
RNase H enzyme (Life Technologies/Invitrogen).
-
10.
Random hexamers (Life Technologies/Invitrogen).
-
11.
N gene primers (see Note 2 ): NF, 5′-TCTTTTGCCATGGCAAGC-3′; NR, 5′-TTGGGTACCTAAAAGTTCATTCTC-3′.
-
12.
Taq polymerase (Promega Corp, Madison, WI, USA).
-
13.
Pfu polymerase (Stratagene, La Jolla, CA, USA).
-
14.
Deoxynucleotide triphosphates (dNTP) (Promega).
-
15.
Sterile distilled water.
-
16.
pTriEx 1.1 cloning kit (Novagen, Madison, WI, USA; EMD Millipore Corp., Billerica, MA, USA).
-
17.
Vector primer for clone screening: TriExUP primer (Novagen; EMD Millipore Corp.).
-
18.
QIAquick PCR product purification kit (Qiagen, Valencia, CA, USA).
-
19.
Restriction enzymes: Nco I and Kpn I (New England Biolabs, Inc., Ipswich, MA, USA) (see Note 3 ).
-
20.
Shrimp alkaline phosphatase (SAP) (Promega Corp).
-
21.
Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA, USA).
-
22.
Escherichia coli (E. coli) NovaBlue competent cell (Novagen; EMD Millipore Corp.).
-
23.
Growth medium: LB medium containing 50 μg/ml carbenicillin (Life Technologies, Grand Island, NY, USA).
-
24.
QIAquick mini-prep kit (Qiagen).
-
25.
E. coli Tuner (DE3) pLacI competent cell (Novagen; EMD Millipore Corp.).
2.2 Expression of Recombinant TCoV Protein
-
1.
Expression host E. coli cell, strain Tuner (DE3) pLacI transformant with recombinant plasmid pTri-N containing N protein gene (prepared from Sects. 2.1 and 3.1).
-
2.
Expression medium: LB medium containing 50 μg/ml carbenicillin, 34 μg/ml chloramphenicol, and 1 % glucose (Life Technologies) (see Note 4 ).
-
3.
Isopropyl β-d-1-thiogalactopyranoside (IPTG), 100 mM (Life Technologies).
-
4.
Incubator shaker (New Brunswick Scientific Co., Inc., Edison, NJ, USA; Eppendorf North America, Hauppauge, NY, USA).
2.3 Purification of Recombinant TCoV N Protein
-
1.
BugBuster (Novagen; EMD Millipore Corp.).
-
2.
Benzonase (Novagen; EMD Millipore Corp.).
-
3.
His-Bind column, pre-packed column with 1.25 ml Ni2+ charged His-Bind resin (Novagen; EMD Millipore Corp.) (see Note 5 ).
-
4.
Binding buffer, 5 mM imidazole, 0.5 M NaCl, 20 mM Tris–HCl (pH 7.9) (Novagen; EMD Millipore Corp.) containing 6 M urea (see Note 6 ).
-
5.
Wash buffer, 20 mM imidazole, 0.5 M NaCl, 20 mM Tris–HCl (pH 7.9) (Novagen; EMD Millipore Corp.) containing 6 M urea.
-
6.
Elute buffer, 1 M imidazole, 0.5 M NaCl, 20 mM Tris–HCl (pH 7.9) (Novagen; EMD Millipore Corp.) containing 6 M urea.
-
7.
Protein assay reagent (Bio-Rad, Hercules, CA, USA).
-
8.
SDS-polyacrylamide gel electrophoresis and Western blotting apparatuses (Mini-Protean® electrophoresis chamber and wet/tank blotting system, Bio-Rad).
3 Methods
3.1 Construction of TCoV N Gene in the Expression Vector pTriEx
-
1.
Total RNA was extracted from TCoV virus source by RNApure reagent. Two hundred microliters of virus solution are mixed with 1 ml of RNApure reagent and incubated on ice for 10 min (see Note 7 ).
-
2.
Add 180 μl of chloroform, mix the mixture, and vortex vigorously for 10 s (see Note 8 ).
-
3.
Centrifuge at 13,000 × g for 10 min at 4 °C. Carefully take the upper aqueous phase into a clean microcentrifuge tube and mix with equal volume of cold isopropanol by vortexing vigorously for 30 s. Incubate on ice for 10 min.
-
4.
Centrifuge at 13,000 × g for 10 min at 4 °C. Carefully discard the supernatant without disturbing the RNA pellet.
-
5.
Wash RNA pellet with 1 ml of cold 70 % ethanol. Incubate on ice for 5 min.
-
6.
Centrifuge at 13,000 × g for 2 min at 4 °C. Remove ethanol. Spin briefly and remove the residual liquid with pipette (see Note 9 ).
-
7.
Dissolve RNA pellet in 50 μl of DEPC-H2O and a portion of it is quantified by spectrophotometry (GeneQuant Pro Spectrophotometer, Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA; GeneQuant 1300 Spectrophotometer, GE Healthcare Bio-Sciences, Piscataway, NJ, USA) at 260 nm wavelength (see Note 10 ).
-
8.
Mix 8 μl (1 pg to 5 μg) of RNA with 1 μl (50 ng/μl) random hexamer and 1 μl (10 mM) dNTP in a total volume of 10 μl.
-
9.
Incubate at 65 °C for 5 min and sit on ice for 1 min.
-
10.
Add 10 μl of SuperScript III cDNA Synthesis Mix (containing 2 μl 10× RT buffer, 4 μl (25 mM) MgCl2, 2 μl (0.1 M) DTT, 1 μl (40 U/μl) RNaseOUT, 1 μl (200 U/μl) SuperScript III RT enzyme) to each RNA/primer mixture.
-
11.
Incubate at 25 °C for 10 min followed by 50 °C for 50 min.
-
12.
Terminate the reverse transcription (RT) reaction at 85 °C for 5 min and chill on ice (see Note 11 ).
-
13.
Add 2 μl of the above RT mixture to the PCR amplification reaction (100 μl) with primers NF and NR. A mix of Taq and Pfu at 10:1 is recommended to maintain PCR fidelity (Table 1).
Table 1 Preparation of reaction mixture for turkey coronavirus nucleocapsid protein gene polymerase chain reaction -
14.
PCR cyclic parameters: 94 °C for 10 s for denaturation, 58 °C for 30 s for annealing, 72 °C for 2 min for extension for 35 cycles followed by 72 °C for 10-min final extension.
-
15.
PCR product is purified by QIAquick PCR purification kit and recovered in 16 μl of sterile distilled water (pH 7.0–8.5).
-
16.
Add all 16 μl of purified PCR product to the restriction enzyme (RE) digestion with 1 μl (10 units/μl) Nco I, 1 μl (10 units/μl) Kpn I, and 2 μl 10× buffer in one reaction of 20 μl. Incubate at 37 °C for 4 h.
-
17.
Set up the same RE digestion for 1 μg of vector pTriEx 1.1 vector plasmid.
-
18.
For dephosphorylation of digested vector, add 1 μl (1 unit) of SAP to the 20 μl of digestion reaction mixture. Incubate at 37 °C for 30 min.
-
19.
Gel purify both digestion reactions for PCR product and plasmid pTriEx by Zymoclean gel DNA recovery kit. Recover each digested DNA prep in 10 μl of sterile distilled water (see Note 12 ).
-
20.
Set up two ligation reactions (pTriEx 1.1 cloning kit) with 1 μl of pTriEx vector and 1 or 4 μl of PCR product (insert) DNA in a reaction of 10 μl as demonstrated in Table 2. Incubate at 16 °C for 15 min (see Note 13 ).
Table 2 Preparation of ligation reaction mixture for recombinant plasmid carrying turkey coronavirus nucleocapsid protein gene -
21.
Take 1 μl of ligation reaction for transformation to NovaBlue competent cell. Plate 20 and 100 μl of the transformation mix on growth medium plates (see Note 14 ).
-
22.
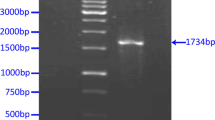
Colony screening by PCR with primers TriExUp and NR. The PCR cycling parameters are the same as above for N gene amplification (step 14). The PCR product of correct clone is about 1500 bp.
-
23.
The selected clone is grown in growth medium. Plasmids are purified by QIAquick mini-prep kit and sequenced to confirm that the inserted TCoV N gene is in frame with the vector-defined open reading frame at both its N- and C-termini.
-
24.
The recombinant plasmid containing the entire TCoV N protein gene (pTri-N) is transformed to competent E. coli strain Tuner (DE3) pLacI. Transformants are grown in expression medium.
3.2 Expression of Recombinant TCoV N Protein
-
1.
A starter culture of expression host bacteria cells is prepared in 3 ml of expression medium. Cells are incubated at 37 °C with shaking at 250 rpm in an incubator shaker overnight, to an OD of 600 nm approximately 0.5 (see Note 15 ).
-
2.
The entire 3 ml culture is added to 100 ml of fresh expression medium and incubated at 37 °C with shaking at 250 rpm in an incubator shaker until the OD reached 0.5–1.0 at 600 nm wavelength (see Note 16 ).
-
3.
The cultures are induced by addition of IPTG to a final concentration of 1 mM (add 1 ml of sterile 100 mM IPTG solution). The induced cultures are incubated at 37 °C with shaking at 250 rpm in an incubator shaker for another 4 h.
-
4.
The bacterial cultures are harvested by centrifugation at 10,000 × g for 10 min at 4 °C. The cell pellet can be immediately processed or frozen stored at −20 or −80 °C until processed (see Note 17 ).
3.3 Purification of Recombinant TCoV N Protein
-
1.
Resuspend the cell pellet completely in 5 ml of BugBuster reagent per gram of wet cell weight (see Note 18 ).
-
2.
The nuclease reagent Benzonase is added with 1 μl (25 units) for every ml of BugBuster reagent used.
-
3.
Incubate at room temperature with slow rotation for 20 min.
-
4.
Centrifuge at 16,000 × g for 20 min at 4 °C. The soluble supernatant is discarded and the insoluble fraction (containing inclusion bodies) is dissolved in 10 ml of binding buffer (see Note 19 ).
-
5.
Equilibrate His-Bind column with 10 ml of binding buffer. Allow the entire buffer volume to flow through.
-
6.
Apply the protein solution (dissolved inclusion bodies) to the column.
-
7.
Wash the column with 10 ml of binding buffer.
-
8.
Wash the column with 10 ml of wash buffer.
-
9.
Elute the recombinant N protein from the column with 5 ml of elute buffer (see Note 20 ).
-
10.
The concentration of purified N protein is determined by protein assay reagent. The yield of purified N protein is about 10 mg from a 100 ml of culture (see Note 21 ).
-
11.
The purified N protein is further confirmed by SDS-polyacrylamide gel electrophoresis and Western blotting as a single protein band with a molecular mass of about 57 kDa.
4 Notes
-
1.
Homogenates of infected intestines can be the virus source. For better results, further purification of intestinal homogenates containing TCoV with 40–60 % sucrose gradient is recommended. TCoV/IN/94 (GenBank accession number EU022525) has been used as the stock virus for expression of recombinant TCoV N protein in our laboratory.
-
2.
Primers NF and NR containing restriction sites Nco I and Kpn I, respectively. The amplified product containing the entire open reading frame of TCoV N protein gene (1230 bp).
-
3.
Nco I and Kpn I are typical restriction enzymes. The digestions by both enzymes can be conducted in a single reaction with compatible buffer. For example, the two enzymes from New England Biolabs (Ipswich, MA) can be performed in NE Buffer 1.1.
-
4.
Ampicillin antibiotic marker is on the expression vector pTriEx and chloramphenicol antibiotic marker is on plasmid pLacI in the expression host strain Tuner cells. Carbenicillin is recommended to be in place of ampicillin for better stability for pH changes throughout the bacterial cultures.
-
5.
The binding capacity of 1.25 ml of His-Bind resin is 10 mg of target protein per column. As for any affinity chromatography, the best purity of target protein is achieved when the amount of protein extract is near the binding capacity.
-
6.
The purpose of 6 M urea is to improve resolution of the sticky inclusion bodies. The presence of 6 M urea does not affect binding of His-Bind resin to target N protein.
-
7.
The suggested ratio of RNApure reagent to sample is 10:1. Excess amount of RNApure reagent has no negative impact. The lower ratio (5:1) in this step is intended to obtain higher concentration of viral RNA in the final supernatants. If the upper aqueous phase after centrifugation at step 3 is more than half of the total volume, there is not enough RNApure reagent added. The appropriate reagent amount may be adjusted. Chloroform is applied at 150 μl for every milliliter of lysate.
-
8.
The sample mixture with chloroform at this step can be stored at −70 °C or even lower temperature before proceeding to the next step.
-
9.
Optional: Inverting the tube for 5–10 min for air-drying of RNA pellet is a helpful tip to completely remove any residual ethanol that may interfere the following RT reaction.
-
10.
It is critical to make sure that the jellylike RNA pellet is completely dissolved into solution by repeat pipetting. The volume (50 μl) of DEPC-H2O may be adjusted according to RNA pellet size to achieve appropriate concentrations. Concentration can be estimated by taking 1 μl of the RNA solution into 1 ml of water. Read at 260 nm. 1 OD260 = 40 μg. The RNA quality can be further examined by OD 260/280 and 260/230 ratio. The ratio of 260/280 about 2.0 is considered as pure for RNA, while 1.8 is considered pure for DNA . The expected 260/230 ratio is around 2.0–2.2 for pure nucleic acid. If the ratio is appreciably lower, it may indicate the presence of protein, phenol, or other contaminants with strong absorption near 280 or 230 nm. RNA should be stored at −70 °C or even lower temperature.
-
11.
The synthesized cDNA in the RT reaction can be stored at −20 °C or even lower temperature until used.
-
12.
PCR product may be purified. The vector must be gel purified due to the long digested fragment size above 30 bp.
-
13.
The molar ratio between vector and insert is suggested at 1:2 to 1:5. The volumes in this step are illustrated for initial exploration. The concentration of digested vector and insert can be estimated by OD 260 or agarose gel electrophoresis with known amount of DNA of similar size in adjacent wells. The ligation reaction mixture can be stored at 4 °C until used for transformation or at −20 °C for longer term.
-
14.
After plating, the leftover transformation mix can be stored at 4 °C for further plating in the following days at different amount if needed.
-
15.
The starter culture can be prepared from a fresh colony on a plate or directly from a glycerol storage stock. An OD around 0.5 represents a culture at log phase when the cells are at the best condition to expand and for protein expression.
-
16.
This usually takes about 2–3 h to reach the OD range. The higher the OD of starter culture in the previous step, the shorter the time to reach this OD range.
-
17.
The centrifuge tubes should be weighed before and after collection of cell pellet for estimation of wet pellet amount and the volume of BugBuster to be applied in the next step. Frozen storage of cell pellets may improve the extraction efficiency of BugBuster reagents through the freeze/thaw cycle.
-
18.
It is important to completely resuspend the cell pellets for the best results of BugBuster extraction. Higher volume of BugBuster reagent does not have adverse effect. Roughly 10–20 ml of BugBuster reagent should be enough for cell pellets collected from a 100 ml of culture. BugBuster reagent can be added directly to frozen cell pellets. There is no need to wait for the temperature to return to room temperature. Protease inhibitors may be added at this step but usually not necessary.
-
19.
It is critical but somewhat difficult to completely dissolve the sticky inclusion bodies. Repeat pipetting up and down until the protein solution is homogeneous. Any undissolved particles will clot the His-Bind column and affect the purification process. It is advisable to centrifuge the dissolved inclusion body protein solution at 5000–10,000 × g for 10–15 min at 4 °C for clarification before application to the column.
-
20.
Eluate may be collected in fractions such as 0.5 or 1 ml each fraction.
-
21.
The presence of 6 M urea is compatible with the protein assay reagent. The assay range can be adjusted for protein concentrations from low μg/ml to 1 mg/ml with different assay format. The protein concentration of the target N protein eluate as obtained following this process is about 1–2 mg/ml. The presence of 6 M urea has no adverse effect on plate coating for ELISA performance. Given the coating concentration of N protein at 20 μg/ml, the eluate is usually diluted in coating buffer for at least 1:10 to reduce the urea content to less than 600 mM and, subsequently, further diminish any possible effect on ELISA performance. Accordingly, the purified N protein eluate can be directly applied to the ELISA assay for detection of antibodies to TCoV.
References
Nagaraja KV, Pomeroy BS (1997) Coronaviral enteritis of turkeys (blue comb disease). In: Calnek BW, Barnes HJ, Beard CW et al (eds) Diseases of poultry, 10th edn. Iowa state University Press, Ames, IA
Pomeroy BS, Larsen TC, Deshmukh RD, Patel LB (1975) Immunity to transmissible coronaviral enteritis of turkeys (Blue comb). Am J Vet Res 36:553–555
Dea S, Tijssen P (1988) Identification of the structural proteins of turkey enteric coronavirus. Arch Virol 99:173–186
Saif LJ (1993) Coronavirus immunogens. Vet Microbiol 37:285–297
Linde GA, Granstrom M, Orvell C (1987) Immunoglobulin class and immunoglobulin G subclass enzyme-linked immunosorbent assays compared with microneutralisation assay for sero-diagnosis of mumps infection and determination of immunity. J Clin Microbiol 25:1653–1658
Reid-Sanden FL, Sumner JW, Smith JS, Fekadu M, Shaddock JH, Bellini WJ (1990) Rabies diagnostic reagents prepared from a rabies N gene recombinant expressed in baculovirus. J Clin Microbiol 28:858–863
Hummel KB, Erdman DD, Heath J, Bellini WJ (1992) Baculovirus expression of the nucleocapsid gene of measles virus and utility of the recombinant protein in diagnostic enzyme immunoassays. J Clin Microbiol 30:2874–2880
Ahmad S, Bassiri M, Banerjee AK, Yilma T (1993) Immunological characterization of the VSV nucleocapsid (N) protein expressed by recombinant baculovirus in Spodoptera exigua larva: use in differential diagnosis between vaccinated and infected animals. Virology 192:207–216
Errington W, Steward M, Emmerson P (1995) A diagnostic immunoassay for Newcastle disease virus based on the nucleocapsid protein expressed by a recombinant baculovirus. J Virol Methods 55:357–365
Ndifuna A, Waters AK, Zhou M, Collisson EW (1998) Recombinant nucleocapsid protein is potentially an inexpensive, effective serodiagnostic reagent for infectious bronchitis virus. J Virol Methods 70:37–44
Loa CC, Lin TL, Wu CC, Bryan TA, Hooper T, Schrader D (2004) Expression and purification of turkey coronavirus nucleocapsid protein in Escheria coli. J Virol Methods 116:161–167
Abdelwahab M, Loa CC, Wu CC, Lin TL (2015) Recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay for detection of antibody to turkey coronavirus. J Virol Methods 217:36–41
Acknowledgements
The protocol “Recombinant turkey coronavirus nucleocapsid protein expressed in Escherichia coli” detailed in this chapter had been successfully carried out in the authors’ studies on characterization and immunology of turkey coronaviral enteritis . Those studies were in part financially supported by USDA, North Carolina Poultry Federation, and/or Indiana Department of Agriculture and technically assisted by Drs. Tom Brien and David Hermes, Mr. Tom Hooper, and Ms. Donna Schrader for clinical and diagnostic investigation, virus isolation and propagation, and animal experimentation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Loa, C.C., Wu, C.C., Lin, T.L. (2016). Recombinant Turkey Coronavirus Nucleocapsid Protein Expressed in Escherichia coli . In: Wang, L. (eds) Animal Coronaviruses. Springer Protocols Handbooks. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3414-0_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3414-0_4
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3412-6
Online ISBN: 978-1-4939-3414-0
eBook Packages: Springer Protocols