Abstract
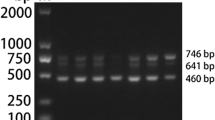
A multiplex polymerase chain reaction (PCR) method for differential detection of turkey coronavirus (TCoV), infectious bronchitis virus (IBV), and bovine coronavirus (BCoV) is presented in this chapter. Primers are designed from the conserved or variable regions of nucleocapsid (N) or spike (S) protein genes of TCoV, IBV, and BCoV and used in the same PCR reaction. Reverse transcription followed by PCR reaction is used to amplify a portion of N or S gene of the corresponding coronaviruses. Two PCR products, a 356-bp band corresponding to N gene and a 727-bp band corresponding to S gene, are obtained for TCoV. In contrast, one PCR product of 356 bp corresponding to a fragment of N gene is obtained for IBV strains and one PCR product of 568 bp corresponding to a fragment of S gene is obtained for BCoV.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Turkey coronavirus (TCoV) contributed to significant economic losses and remains as a serious threat to the turkey producers. Turkey coronaviral enteritis in areas with high concentrations of turkeys on a year-round basis is not easily eliminated and is encountered frequently in turkey poults [1]. Accurate and rapid method for diagnosis of TCoV infection is the key to effective control of the disease.
Turkey coronavirus belongs to the family Coronaviridae, which is a group of enveloped, positive-stranded RNA viruses that infect a wide range of mammalian and avian species. The major structural proteins of coronavirus include phosphorylated nucleocapsid (N) protein, peplomeric spike (S) glycoprotein, and transmembrane or membrane (M) glycoprotein. Spike protein contributes to the distinctive peplomers on the viral surface and contains neutralizing and group-specific epitopes. Spike protein is highly variable among different coronaviruses while M and N proteins are more conserved among coronaviruses between different antigenic groups [2].
There is a close antigenic and genomic relationship between TCoV and infectious bronchitis virus (IBV) according to studies of immunofluorescent antibody assay (IFA ), enzyme-linked immunosorbent assay (ELISA ), and sequence analysis in our and other laboratories [3–8]. In addition, bovine coronavirus (BCoV) was demonstrated to cause experimental enteric infection in turkey [9]. Therefore, close relationship between TCoV and BCoV was previously reported and TCoV was placed in an antigenic group as BCoV [10, 11]. Although the sequence data revealed divergence of S genes among TCoV, IBV, and BCoV [12–14], there is still a need to detect and differentiate them accurately and quickly. Polymerase chain reaction (PCR ) assay has been an important approach for detecting many veterinary important microorganisms with the distinct advantages of high sensitivity and specificity. This chapter describes a multiplex PCR assay to detect and differentiate TCoV, IBV, and BCoV in a single reaction [15].
2 Materials
2.1 Viral RNA Preparation
-
1.
Test sample: Turkey intestine .
-
2.
Phosphate-buffered saline (PBS), pH 7.4.
-
3.
Stomacher® 80 microBiomaster laboratory paddle blenders (Seward, Davie, FL, USA).
-
4.
RNApure reagent (GenHunter, Nashville, TN, USA).
-
5.
Chloroform.
-
6.
Isopropanol.
-
7.
70 % Ethanol.
-
8.
Diethyl-pyrocarbonate (DEPC)-treated sterile double-distilled water (DEPC-H2O).
2.2 Reverse Transcription
-
1.
SuperScript III first-strand synthesis system for RT-PCR kit (Life Technologies/Invitrogen, Carlsbad, CA, USA).
-
2.
RNaseOUT is a recombinant RNase inhibitor (Life Technologies/Invitrogen).
-
3.
Random hexamers (Life Technologies/Invitrogen).
-
4.
Deoxynucleotide triphosphates (dNTP) (Promega Corp., Madison, WI, USA).
2.3 PCR Reaction
-
1.
Primers listed in Table 1 (see Note 1 ).
Table 1 List of primers and sequences for coronavirus multiplex polymerase chain reaction -
2.
Taq polymerase (Promega Corp).
-
3.
Sterile distilled water.
-
4.
PCR thermal cycler (GeneAmp PCR System 9600, Perkin Elmer Centers Corp., Norwalk, CT, USA).
-
5.
Agarose.
-
6.
Ethidium bromide, 0.5 μg/ml.
-
7.
Ultraviolet transilluminator.
3 Methods
3.1 Viral RNA Preparation
-
1.
Turkey intestines are cut into pieces and homogenized by Stomacher with fivefold volume of PBS solution.
-
2.
The intestinal homogenates are clarified by centrifugation at 1500 × g for 10 min. The supernatants containing TCoV are used as virus source for preparation of RNA templates for reverse transcription (RT)-PCR reaction.
-
3.
Two hundred microliters of above supernatants are mixed with 1 ml of RNApure reagent and incubated on ice for 10 min (see Note 2 ).
-
4.
Add 180 μl of chloroform, mix the mixture, and vortex vigorously for 10 s (see Note 3 ).
-
5.
Centrifuge at 13,000 × g for 10 min at 4 °C. Carefully take the upper aqueous phase into a clean tube and mix with equal volume of cold isopropanol by vortexing vigorously for 30 s. Incubate on ice for 10 min.
-
6.
Centrifuge at 13,000 × g for 10 min at 4 °C. Carefully discard the supernatant without disturbing RNA pellet.
-
7.
Wash RNA pellet with 1 ml of cold 70 % ethanol. Incubate on ice for 5 min.
-
8.
Centrifuge at 13,000 × g for 2 min at 4 °C. Remove the ethanol. Spin briefly and remove the residual liquid with pipette (see Note 4 ).
-
9.
Dissolve RNA pellet in 50 μl of DEPC-H2O and a portion of it is quantified by spectrophotometry (GeneQuant Pro Spectrophotometer, Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA; GeneQuant 1300 Spectrophotometer, GE Healthcare Bio-Sciences, Piscataway, NJ, USA) at 260 nm wavelength (see Note 5 ).
-
10.
Positive control samples including turkey intestinal homogenates containing TCoV/IN/540/94 (GenBank accession number EU022525), IBV strains (ATCC, Manassas, VA, USA), and BCoV (NVSL, Ames, IA, USA) are processed for viral RNA following the procedures above.
3.2 Reverse Transcription
-
1.
Mix 8 μl (1 pg to 5 μg) of RNA with 1 μl (50 ng/μl) random hexamer and 1 μl (10 mM) dNTP in a total volume of 10 μl.
-
2.
Incubate at 65 °C for 5 min and sit on ice for 1 min.
-
3.
Add 10 μl of SuperScript III cDNA Synthesis Mix (containing 2 μl 10× RT buffer, 4 μl (25 mM) MgCl2, 2 μl (0.1 M) DTT, 1 μl (40 U/μl) RNaseOUT, 1 μl (200 U/μl) SuperScript III RT enzyme) to each RNA/primer mixture.
-
4.
Incubate at 25 °C for 10 min and followed by 50 °C for 50 min.
-
5.
Terminate the RT reaction at 85 °C for 5 min and chill on ice (see Note 6 ).
3.3 PCR Reaction
-
1.
Two microliters of cDNA are used in PCR amplifications. The components for 100 μl reaction mixture are listed in Table 2 (see Note 7 ).
Table 2 Preparation of reaction mixture for coronavirus multiplex polymerase chain reaction -
2.
PCR cyclic parameters: 94 °C for 30 s for denaturation, 50 °C for 1 min for annealing, 72 °C for 1 min for extension for 25 cycles followed by 72 °C for 10-min final extension.
-
3.
A volume of 8 μl of the amplified PCR products is subjected to electrophoresis at 100 V in horizontal gels containing 1 % agarose. The gel is stained with ethidium bromide. Amplified DNA products are visualized by viewing the gel with an ultraviolet transilluminator, compared with standard markers of DNA size (see Notes 8 and 9 ).
4 Notes
-
1.
Primers are based on the variable and conserved regions of S and N gene sequences among TCoV, IBV, and BCoV. The sense primer N103F and antisense primer N102R common to both TCoV and IBV were designed according to the conserved regions of N gene sequences. This set of primers specified a 356-bp sequence corresponding to nucleotide position 445 to 801 of TCoV N gene. The sense primer S306F and antisense primer S306R specific to TCoV were designed according to the variable regions of S gene sequences among these viruses. This set of primers specified a 727-bp sequence corresponding to the nucleotide position 2019 to 2745 of TCoV S gene. The sense primer S3 and antisense primer S6 specific to BCoV were designed according to the variable regions of S gene sequences among these viruses. This set of primers specified a 568-bp sequence corresponding to the nucleotide position 1488 to 2055 of BCoV S gene.
-
2.
The suggested ratio of RNApure reagent to sample is 10:1. Excess amount of RNApure reagent has no negative impact. The lower ratio (5:1) in this step is intended to obtain higher concentration of viral RNA in the final supernatants. If the upper aqueous phase after centrifugation at step 5 is more than half of the total volume, there is not enough RNApure reagent added. The appropriate reagent amount may be adjusted. Chloroform is applied at 150 μl for every ml of lysate.
-
3.
The sample mixture with chloroform at this step can be stored at −70 °C or lower than −70 °C before proceeding to the next step.
-
4.
Optional: Invert the tube for 5–10 min for air-drying of RNA pellet is a helpful tip to completely remove any residual ethanol that may interfere with the following RT reaction.
-
5.
It is critical to make sure that the jellylike RNA pellet is completely dissolved into solution by repeat pipetting. The volume (50 μl) of DEPC-H2O may be adjusted according to RNA pellet size for appropriate RNA concentration. Concentration can be estimated by taking 1 μl of the RNA solution into 1 ml of water. Read at 260 nm. 1 OD260 = 40 μg. The RNA quality can be further examined by OD 260/280 and 260/230 ratio. The ratio of 260/280 about 2.0 is considered as pure for RNA, while 1.8 is considered pure for DNA . The expected 260/230 ratio is around 2.0 to 2.2 for pure nucleic acid. If the ratio is appreciably lower, it may indicate the presence of protein, phenol, or other contaminants with strong absorption near 280 or 230 nm. RNA should be stored at −70 °C or lower than −70 °C.
-
6.
The synthesized cDNA in the RT reaction can be stored at −20 °C or lower than −20 °C until used. RNase H digestion to remove the RNA templates from the cDNA:RNA hybrid molecule is optional. The RNase treatment may increase PCR sensitivity for long templates. The target products (356, 568, and 727 bp) of this multiplex PCR are not considered long. The benefit of this RNase digestion is likely limited.
-
7.
For routine practice in the laboratory for large number of samples, 25 μl reaction can be applied with reduced cost of reagents.
-
8.
Result interpretation:
-
TCoV positive: two PCR products, a 356-bp band, and a 727-bp band.
-
IBV positive: one PCR product of 356 bp.
-
BCoV positive: one PCR product of 568 bp.
-
Negative: no PCR product.
-
One limitation of this multiplex PCR should be noted. When both TCoV and IBV are present in the sample, the IBV-positive PCR product at 356 bp is overlapping with one of the two TCoV PCR products. The result of two PCR product bands at 356 and 727 bp reflects a confirmed diagnosis of TCoV positive but does not rule out the presence of IBV. This concern is alleviated by tissue tropism of IBV. Turkey enteric infection by chicken respiratory IBV has never been reported. Accordingly, the presence of IBV in turkey intestines (the test sample) is not likely. Any such concern should be further evaluated by a separate PCR specific for IBV [16]. On the other hand, IBV-positive result is able to exclude the presence of TCoV.
-
-
9.
The sensitivity of this multiplex PCR is 4.8 × 10−3 μg of TCoV RNA, 4.6 × 10−4 μg of IBV RNA, and 8.0 × 10−2 μg of BCoV RNA. The specificity had been evaluated against Newcastle disease virus, Marek’s disease virus, turkey pox virus, pigeon pox virus, fowl pox virus, reovirus, infectious bursal disease virus, enterovirus, astrovirus, Salmonella enterica, Escherichia coli, and Mycoplasma gallisepticum with negative results that have no PCR products.
References
Nagaraja KV, Pomeroy BS (1997) Coronaviral enteritis of turkeys (blue comb disease). In: Calnek BW, Barnes HJ, Beard CW et al (eds) Diseases of poultry, 10th edn. Iowa state University Press, Ames, IA
Saif LJ (1993) Coronavirus immunogens. Vet Microbiol 37:285–297
Guy JS, Barnes HJ, Smith LG, Breslin J (1997) Antigenic characterization of a turkey coronavirus identified in poult enteritis and mortality syndrome-affected turkeys. Avian Dis 41:583–590
Stephensen CB, Casebolt DB, Gangopadhyay NN (1999) Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res 60:181–189
Breslin JJ, Smith LG, Fuller FJ, Guy JS (1999) Sequence analysis of the matrix/nucleocapsid gene region of turkey coronavirus. Intervirology 42:22–29
Breslin JJ, Smith LG, Fuller FJ, Guy JS (1999) Sequence analysis of the turkey coronavirus nucleocapsid protein gene and 3′ untranslated region identifies the virus as a close relative of infectious bronchitis virus. Virus Res 65:187–193
Loa CC, Lin TL, Wu CC, Bryan TA, Thacker HL, Hooper T, Schrader D (2000) Detection of antibody to turkey coronavirus by antibody-capture enzyme-linked immunosorbent assay utilizing infectious bronchitis virus antigen. Avian Dis 44:498–506
Akin A, Lin TL, Wu CC, Bryan TA, Hooper T, Schrader D (2001) Nucleocapsid protein gene sequence analysis reveals close genomic relationship between turkey coronavirus and avian infectious bronchitis virus. Acta Virol 45(1):31–38
Ismail MM, Cho KO, Ward LA, Saif LJ, Saif YM (2001) Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis 45:157–163
Dea S, Verbeek AJ, Tijssen P (1990) Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J Virol 64:3112–3118
Verbeek A, Tijssen P (1991) Sequence analysis of the turkey enteric coronavirus nucleocapsid and membrane protein genes: a close genomic relationship with bovine coronavirus. J Gen Virol 72:1659–1666
Lin TL, Loa CC, Wu CC (2004) Complete sequences of 3′end coding region for structural protein genes of turkey coronavirus. Virus Res 106(1):61–70
Loa CC, Lin TL, Wu CC, Bryan TA, Hooper T, Schrader D (2006) Comparison of 3′ end encoding regions of turkey coronavirus isolates from Indiana, North Carolina, and Minnesota. Intervirology 49(4):230–238
Cao J, Wu CC, Lin TL (2008) Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res 136(1-2):43–49
Loa CC, Lin TL, Wu CC, Bryan TA, Hooper T, Schrader D (2006) Differential detection of turkey coronavirus, infectious bronchitis virus, and bovine coronavirus by a multiplex polymerase chain reaction. J Virol Methods 131:86–91
Lee CW, Hilt DA, Jackwood MW (2000) Redesign of primers and application of the reverse transcriptase-polymerase chain reaction and restriction fragment length polymorphism test to the DE072 strain of infectious bronchitis virus. Avian Dis 44:650–654
Acknowledgements
The protocol “A multiplex polymerase chain reaction for differential detection of turkey coronavirus from chicken infectious bronchitis virus and bovine coronavirus” outlined in this chapter had been successfully carried out in the authors’ studies on molecular diagnostics and molecular virology of turkey coronavirus infection in turkeys. Those studies were in part financially supported by USDA, North Carolina Poultry Federation, and/or Indiana Department of Agriculture and technically assisted by Drs. Tom Brien and David Hermes, Mr. Tom Hooper, and Ms. Donna Schrader for clinical and diagnostic investigation, virus isolation and propagation, and animal experimentation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Loa, C.C., Wu, C.C., Lin, T.L. (2016). A Multiplex Polymerase Chain Reaction for Differential Detection of Turkey Coronavirus from Chicken Infectious Bronchitis Virus and Bovine Coronavirus. In: Wang, L. (eds) Animal Coronaviruses. Springer Protocols Handbooks. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3414-0_12
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3414-0_12
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3412-6
Online ISBN: 978-1-4939-3414-0
eBook Packages: Springer Protocols