Abstract
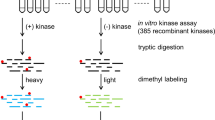
Protein phosphorylation plays an essential role in the regulation of various cellular functions. Dysregulation of phosphorylation is implicated in the pathogenesis of certain cancers, diabetes, cardiovascular diseases, and central nervous system disorders. As a result, protein kinases have become potential drug targets for treating a wide variety of diseases. Identification of kinase substrates is vital not only for dissecting signaling pathways, but also for understanding disease pathologies and identifying novel therapeutic targets. However, identification of bona fide kinase substrates has remained challenging, necessitating the development of new methods and techniques. The kinase assay linked phosphoproteomics (KALIP) approach integrates in vitro kinase assays with global phosphoproteomics experiments to identify the direct substrates of protein kinases. This strategy has demonstrated outstanding sensitivity and a low false-positive rate for kinase substrate screening.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pawson T (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116:191–203
Hunter T (2000) Signaling—2000 and beyond. Cell 100:113–127
Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411:355–365
Cohen P (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem 268:5001–5010
Manning BD, Cantley LC (2002) Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE 162:pe49
Wu RH, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, Gygi SP (2011) A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods 8:677–U111
Jin LL, Tong JF, Prakash A, Peterman SM, St-Germain JR, Taylor P, Trudel S, Moran MF (2010) Measurement of Protein Phosphorylation Stoichiometry by Selected Reaction Monitoring Mass Spectrometry. J Proteome Res 9:2752–2761
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207
Mumby M, Brekken D (2005) Phosphoproteomics: new insights into cellular signaling. Genome Biol 6:230
Altelaar AF, Munoz J, Heck AJ (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet 14:35–48
Cohen P (2002) Protein kinases–the major drug targets of the twenty-first century? Nat Rev Drug Discov 1:309–315
Schwartz D, Gygi SP (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23:1391–1398
Xue L, Wang W, Iliuk A, Hu L, Galan JA, Yu S, Hans M, Geahlen RL, Tao WA (2012) Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc Natl Acad Sci U S A 109:5615–5620
Xue L, Geahlen RL, Tao WA (2013) Identification of direct tyrosine kinase substrates based on protein kinase assay-linked phosphoproteomics. Mol Cell Proteomics 12:2969–2980
Acknowledgments
The authors gratefully acknowledge that this work has been funded in part by an NSF CAREER award CHE-0645020 (WAT), and by National Institutes of Health grant GM088317 (WAT).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Xue, L., Arrington, J.V., Tao, W.A. (2016). Identification of Direct Kinase Substrates via Kinase Assay-Linked Phosphoproteomics. In: von Stechow, L. (eds) Phospho-Proteomics. Methods in Molecular Biology, vol 1355. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-3049-4_18
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3049-4_18
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-3048-7
Online ISBN: 978-1-4939-3049-4
eBook Packages: Springer Protocols